INTRODUCTION
Gastric cancer is the fifth most commonly diagnosed cancer worldwide. Helicobacter pylori is a pathogenic bacterium that causes chronic gastro-duodenal disorders, including gastritis, peptic ulcers, and stomach cancer in humans. Eradication of H. pylori can prevent and slow the progression of non-atrophic chronic gastritis to atrophic gastritis, thereby reducing gastric cancer risk (1, 2). This emphasizes the importance of early treatment of H. pylori infections in decreasing gastric cancer rates and associated mortality. The first-line treatment for H. pylori infection, as recommended by the Centers for Disease Control and Prevention (CDC) and various studies, is triple therapy, combining a proton pump inhibitor with clarithromycin and either amoxicillin or metronidazole (3, 4). However, the increasing prevalence of antibiotic-resistant H. pylori strains poses significant challenges to this treatment approach. Geographically, antibiotic resistance has increased in the US from 7% to 14%; Hong Kong, 4.7–10.3%; Japan, 9.1–18.7% (5, 6, 7). The clarithromycin resistance rate in Korea increased sharply from 2.8% in 1994 to 13.8 in 2003 and from 16.7% in 2005 to 38.5% in 2009 (8, 9). These studies indicate that clarithromycin resistance increases with time, and it is necessary to introduce other treatment options.
Quinolone antibiotics, such as moxifloxacin (MOX) and levofloxacin (LVX), have been used worldwide to treat patients not responding to primary treatment (10, 11). Brazil and Italy reported 90 percent curative effects using LVX or MOX (12, 13). However, the use of this wide range of quinolones has led to increased resistance in H. pylori in some areas and a significant reduction in eradication rates in Turkey and Korea (14, 15). High resistance level of LVX has been reported in numerous other countries including Turkey (29.5%), China (26.6%), Portugal (26.3%), Egypt (23.2%), Senegal (15.0%), and Spain (14.5%) (10, 16, 17, 18, 19, 20, 21). In southern Taiwan, resistance to CIP and LVX in H. pylori has significantly increased from 2004 to 2007 in comparison with that observed during 1998–2003, suggesting the need for continuous surveillance of quinolone resistance in H. pylori(22). While these antibiotics have shown promising results in some regions, the growing incidence of quinolone-resistant H. pylori strains has become a global concern. Studies have reported significant variations in resistance rates across different countries, highlighting the need for continuous surveillance and a deeper understanding of the resistance mechanisms.
Quinolones are antibiotics used to treat H. pylori infections by targeting DNA gyrase, an enzyme composed of two subunits, GryA and GyrB. However, widespread mutations in these subunits contribute to high levels of resistance. Widespread point mutations, N87K and D91N, found in the quinolone resistance-determining region (QRDR, from position 71 to 110) of GyrA, are associated with quinolone resistance in H. pylori (9, 23, 24). In this study, we investigate the mutations beyond QRDR of H. pylori gyrA and gyrB, which are associated with the induction of quinolone resistance using clinical isolates from Korean pediatric patients. However, the role of mutations beyond the QRDR in gyrA and gyrB in conferring quinolone resistance remains unclear. This study aims to fill this knowledge gap by conducting an in-depth analysis of mutations found outside the QRDR in gyrA and gyrB, using clinical isolates from Korean pediatric patients.
MATERIALS AND METHODS
Bacteria strains
Helicobacter pylori Y20-A, 92-400, 92-370 and PED-4487GA isolates resistant to CIP, MOX, LVX, and H. pylori 228, 229, 186 and 210 isolates displaying antibiotic susceptibility were selected. These strains were provided by the Fastidious Specialized Pathogen Resources Bank (a member of the National Culture Collection for Pathogens, GNUH), Jinju, Korea. The data regarding antibiotic resistance is shown in Table 1. H. pylori was incubated at 37°C daily in 10% CO2 in Brucella agar containing 10% bovine serum.
Table 1.
MIC of quinolone-susceptible and -resistant H. pylori strains
PCR and DNA sequencing
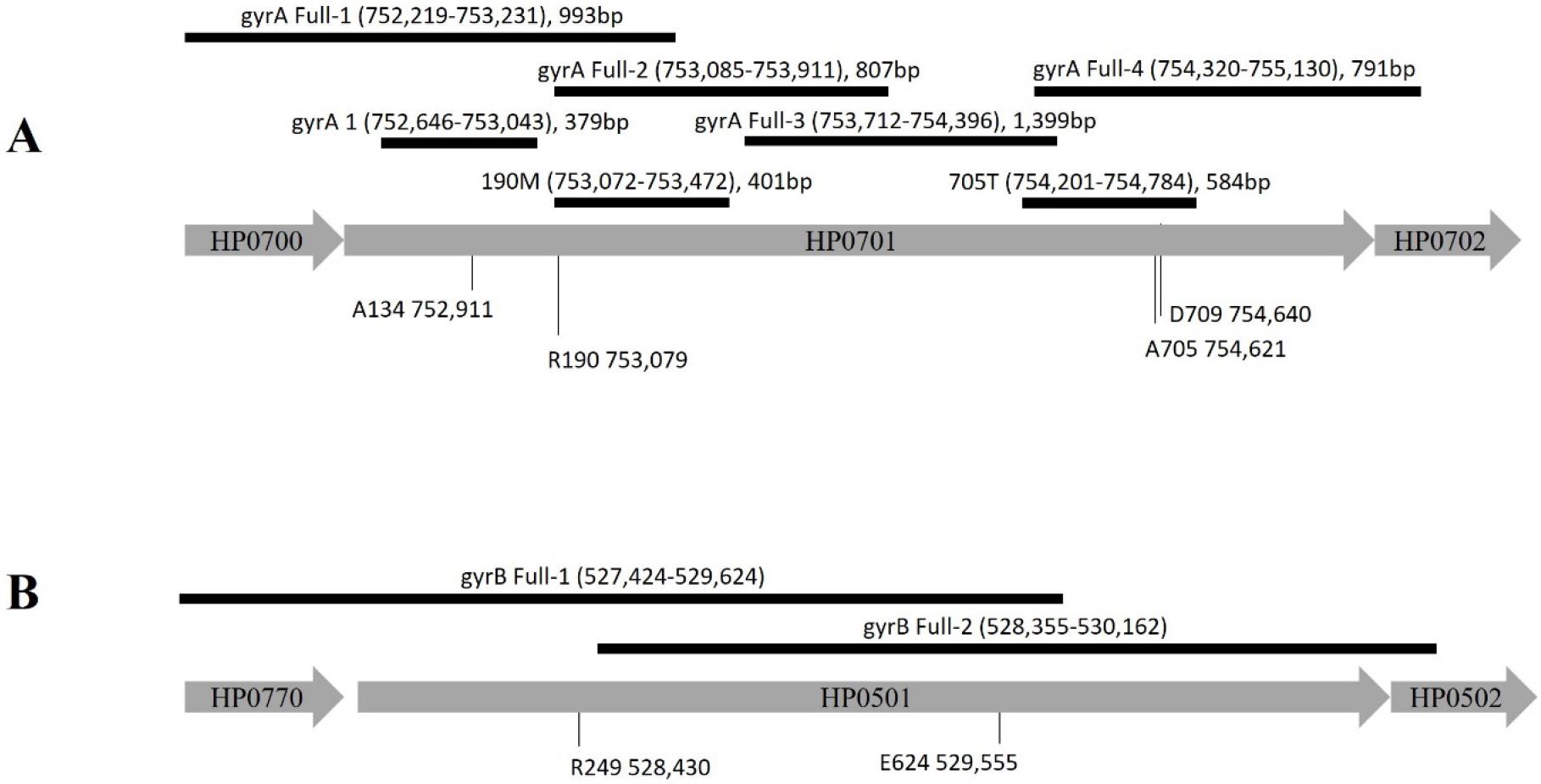
Bacterial gDNA was extracted using the Genomic DNA Extraction Kit (GeNet Bio, Nonsan, Korea) following the manufacturer’s protocols. H. pylori was subjected to PCR and sequencing to analyze the sequence of whole gyrA and the QRDR region of gyrA of the isolates (primers are listed in Table 2, Fig. 1). PCR was performed in 50-μL reaction mixtures using Pfu polymerase (Elpis, Daejeon, Korea). The PCR conditions were as follows: pre-denaturation for 4 min at 94°C, followed by 35 cycles for 1 min at 94°C, 1 min of annealing at 56°C for 30 s, and 1 min at 72°C, followed by final extension for 7 min at 72°C. Amplified DNA was purified using a Favorgen gel/PCR purification kit (Favorgen, Pingtung, Taiwan) for nucleotide sequence analysis following with the manufacturer’s protocol.
Table 2.
Primers of H. pylori gyrA and gyrB for cloning and mutation
| primer | Sequences (5’ → 3’) | Positione | |
|---|---|---|---|
| gyrA 1a |
Forward Reverse |
AAAGCCCGTGCATAGGC TCCCATTAGCCCCATTGA |
752,646 – 753,026 (379 bps) |
| gyrA Full-1b |
Forward Reverse |
GCAAATTGTCATCTTGGCT CCCCGTTTTATAGGCTTCA |
752,219 – 753,213 (993 bps) |
| gyrA Full-2b |
Forward Reverse |
GATGAAATCATAGACGCTTA TTCAAGCGATCTTCGCTCT |
753,085 – 753,893 (807 bps) |
| gyrA Full-3b |
Forward Reverse |
ACAAGCCAAAGCCCAGAAG TGCGTCAAATTGGTGCGC |
753,712 – 755,112 (1,399 bps) |
| gyrA Full-4b |
Forward Reverse |
AAGCACCAAAGACTTTAGCG CTTCGCCCATCACTCTAGA |
754,320 – 755,112 (791 bps) |
| gyrB Full-1c |
Forward Reverse |
GGATTCTGAGCATAGCGAA CGATATAAACATGCCCTTGT |
527,424 – 529,624 (1,808 bps) |
| gyrB Full-2c |
Forward Reverse |
CAATTCGTTAAAGACAGCGC CCACATGCGGTGTCTAAA |
528,355 – 530,162 (1,841 bps) |
| 190Md |
Forward Reverse |
CCCTCACAGGATGGATGAAATC GGTTTAAGACAATTTCACTC |
753,072 – 753,472 (401 bps) |
| 705Te |
Forward Reverse |
AAGGGGCAATTGTATCATTT CTGATAACGCCCACTAGATT |
754,201 – 754,784 (584 bps) |
| A134V upf |
Forward Reverse |
CCACGCGCTTGAATTGCTCT ATGGTTCGCTGGGTTTATCTGGGCGTTAAGAGCGATTTGG |
751,503 – 752,022 (539bp) |
| A134V dof |
Forward Reverse |
TTACTGGATGAATTGTTTTAGTACCACGCCTTGTTAGAAAGCCTTAG GACATGCACTAAAGCGTCTATG |
752,052 – 753,114 (1,088bp) |
| KM |
Forward Reverse |
CCAAATCGCTCTTAACGCCCAGATAAACCCAGCGAACCAT CTAAGGCTTTCTAACAAGGCGTGGTACTAAAACAATTCATCCAGTAA | 1,401bp |
Construction of A134V mutation
The gene of point mutation A134 containing DNA fragment (position 135 to 532) was amplified using a sets of primers (134VDoF and 134VDoR). Upstream of gyrA and the kanamycin resistance gene were amplified by overlapping PCR using two sets of primers (134VupF and 134VupR, KM-F and KM-R). The two PCR products were inserted to pGEM®-5Zf(+) at the PstI restriction site. Resulted constructs were propagated in E. coli grown in LB broth supplemented with ampicillin and kanamycin 50 µg/µl.
Natural transformation
To determine whether point mutations in gyrA are associated with quinolone resistance, genes containing gyrA mutations (gyrA1, 190M and 705T) were transformed into H. pylori 210, 228, and 229 that are susceptible to quinolone. H. pylori, sub-cultured the previous day, was seeded on Brucella agar containing 10% FBS in a loop of approximately 1.5 cm diameter and incubated at 37°C in 10% CO2 for four h. After that, 3 µg of the PCR product containing the gyrA mutant or plasmid clone were supplemented in the H. pylori culture, and the cells were incubated for 24 h at 37°C in 10% CO2. GyrA-transformed H. pylori was selected on Brucella agar plates supplemented with CIP 1 mg/L and 10% bovine serum and cultured for 2–3 d at 37°C in 10% CO2, and colonies were allowed to form. Sequencing analysis was performed, and the minimum inhibitory concentration (MIC) was determined.
Determination of the MIC
The MIC was determined using the agar dilution method (25). Quinolone susceptible H. pylori strain 210 was transformed with the DNA of a resistant strain as a donor. Briefly, overnight-grown H. pylori was resuspended in PBS up to an optical density of 0.5. H. pylori was inoculated in plates containing two-fold dilutions of CIP. Cells were then incubated in 10% CO2 at 37°C for five days. The concentration range of the CIP was 64 mL/L to 0.125 mg/L, and the MIC was determined.
Sequencing analysis
The amino acid sequence was determined through in silico translation using Editseq software (DNASTAR Inc., Madison, WI, USA). Amino acid sequences were aligned using the CLUSTAL X 1.83 program (26) and compared using GeneDoc software (http://www.nrbsc.org/gfx/genedoc).
Molecular docking
GyrA protein sequence was submitted as an input to SWISS-MODEL server (https://www.expasy.org/resources/swiss-model) to run protein structure homology-modelling platform for generating 3D models without assigning any restraints or templates. The highest-quality model was selected based on the Global Model Quality Estimation (GMQE) and Qualitative Model Energy Analysis (QMEAN) scores. Subsequently, the molecular docking was performed using AutoDock Vina to evaluate the binding interactions between GyrA and the CIP. 3D structure of ciprofloxacin (PDB ID: CPF) was downloaded from Protein Data Bank (https://www.rcsb.org/ligand/CPF). The protein and ligand structures were prepared using AutoDockTools. A grid box was defined around the active site to ensure optimal ligand positioning. Docking results were analyzed based on binding affinity scores and molecular interactions.
RESULTS
Detection of gyrA and gyrB mutations in H. pylori clinical isolates
Four quinolone-resistant strains and four quinolone-susceptible strains were randomly selected from H. pylori isolates obtained from Korean children and tested resistance levels by the MIC for quinolone antibiotics CIP, LVX, and MOX (Table 1). MIC values of quinolone antibiotics in the strain 92-370, including CIP (8 mg/L), MOX (2 mg/L), and LVX (4 mg/L), were lower than in the other three quinolone-resistant strains (92-400, Y20-A, and PED-4487GA). Also, QRDR sequences of gyrA of four quinolone-resistant strains were subjected to align with its susceptible strain 210 to mutation analysis. Of the four resistant strains, 92-400, Y20-A, and PED-4487GA harbored mutations at positions 87 and 91 in the QRDR, and N87K and D91N substitutions were observed (Fig. 2A top). On the other hand, no QRDR mutations were observed in both gyrA and gryB of moderated resistant strain 92-370. Therefore, the entire gyrA and gyrB sequences in the strain 92-370 were subjected to determine and compare with these of susceptible strain 210. The strain 92-370 was found to be mutated at eight positions in gyrA, including T5, A134, R190, R398, V492, D594, A705, and N709 (Fig. 2A) and two others in gyrB R249, E624 (Fig. 2B).
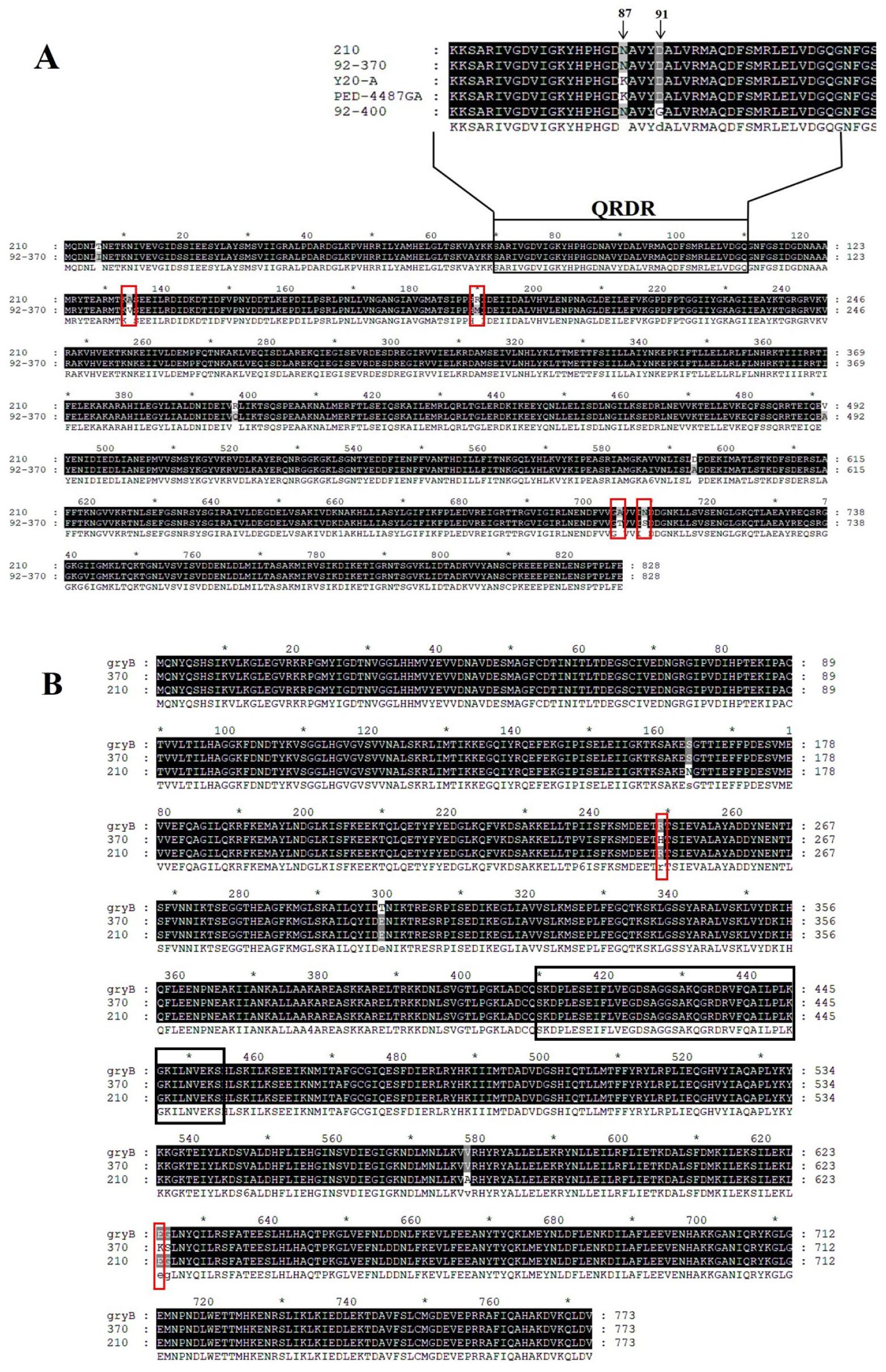
Fig. 2
Sequence alignment of GyrA and GyrB amino acid sequences in susceptible and resistant H. pylori strains. Mutations are indicated in the red box, and the QRDR region is marked by the black box. (A) GyrA sequence alignment between the resistant strain 92-370 and the susceptible strain 210. QRDR region sequence alignment of susceptible strain 210 and resistant strains 92-370, Y20-A, PED-4407GA, and 92-400 is shown on top of the figure. (B) Sequence alignment of GyrB in resistant strain 92-370 (370), susceptible strain 210, and strain 26695 (gyrB).
Confirmation of the association between gyrA and gyrB mutations and quinolone resistance in H. pylori
Among those mutations found in strain 92-370, four mutations have been reported previously: T5, R398, V492, and D594. These mutations were obtained by sequence comparison between 4 fluoroquinolone susceptible strains and 7 resistant strains. Rimbara et al. confirmed that mutations at T5, R398, V492, and D594 were unrelated to quinolone resistance. In this study, to investigate the relation between mutants including A134, R190, A705, and N709 in GyrA and two other R249 and E624 in GyrB and quinolone sensitivity, DNA fragments corresponding to positions containing the mutations mentioned above were natural transformed into the quinolone-susceptible strain 210.
Fig. 3A showed the transformation of DNA fragments containing selected mutations from gyrA and gyrB into susceptible strain 210 (MIC results shown in Table 1). The transformants corresponding to position 3–177, 175–320, and 563–791 of gyrA yielded 87, 12, and 7 colonies in medium supplemented with 1 mg/L CIP, respectively. The transformants of R249H and E624K single mutation inserted DNA fragments of gyrB, which resulted in no colonies. Susceptible strain 210 was also transformed in the same condition simultaneously as a positive control. The results suggested that the A134V mutated strain 210 was highly resistant to CIP 1 mg/L compared to other mutations.
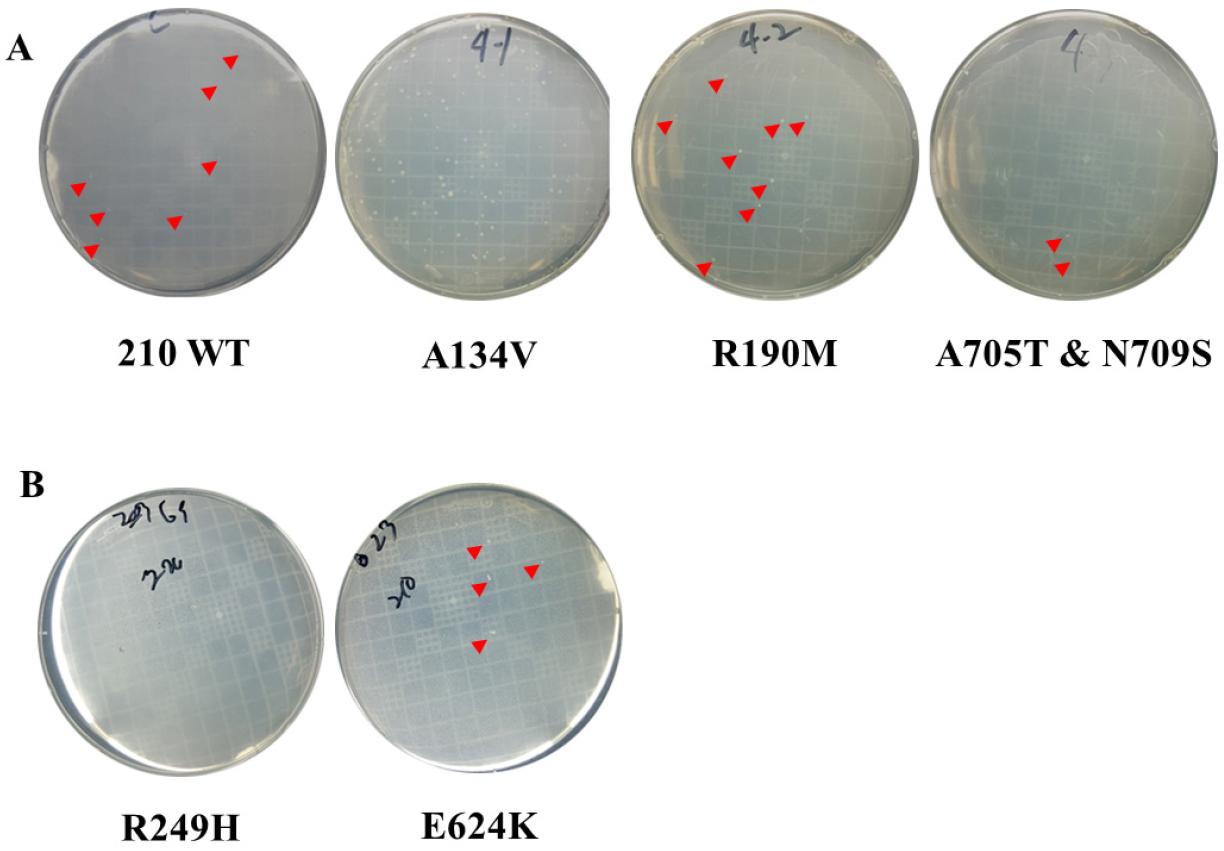
Fig. 3
Confirmation of the relevance of mutations beyond GyrA and GyrB QRDR site resistance to quinolones by natural transformation. (A) Natural transformation to H.pylori strains 210 using each DNA fragment containing A134V, R190M, A705T, and N709S mutations in gyrA. (B) Natural transformation to H.pylori strains 210 using each DNA fragment containing R249H and E624K mutations in gyrB. Red arrowheads indicate bacterial colonies. Due to the high number of colonies on the A134V plate, arrowheads were not added. No colonies were observed for R249H, while R190M, A705T, and N709S produced a few colonies, similar to the 210 WT control.
To evaluate the MIC of CIP among cells transformed with DNA fragments corresponding to DNA fragments 3–177, transformation was performed for quinolone- susceptible strains 210 containing D91G, N87K, and A134V mutations. Four transformants of each mutation were used to determine the MIC of CIP. The susceptible strain 210 was used as the negative control, and the resistant strain 92-370 was used as the positive control. The transformants harboring the D91G and N87K mutations gave a MIC of 32 mg/L for CIP, similar to that of quinolone-resistant clinical strains. However, strains 92-370 yielded an MIC of 2 mg/L for CIP, while transformants with A134V mutants yielded an MIC of 32 mg/L for CIP (Fig. 4A). Sequencing results of those transformants showed that other D91G mutations appeared in the QRDR region along with the A134V mutation in the susceptible strain 210 (data not shown).
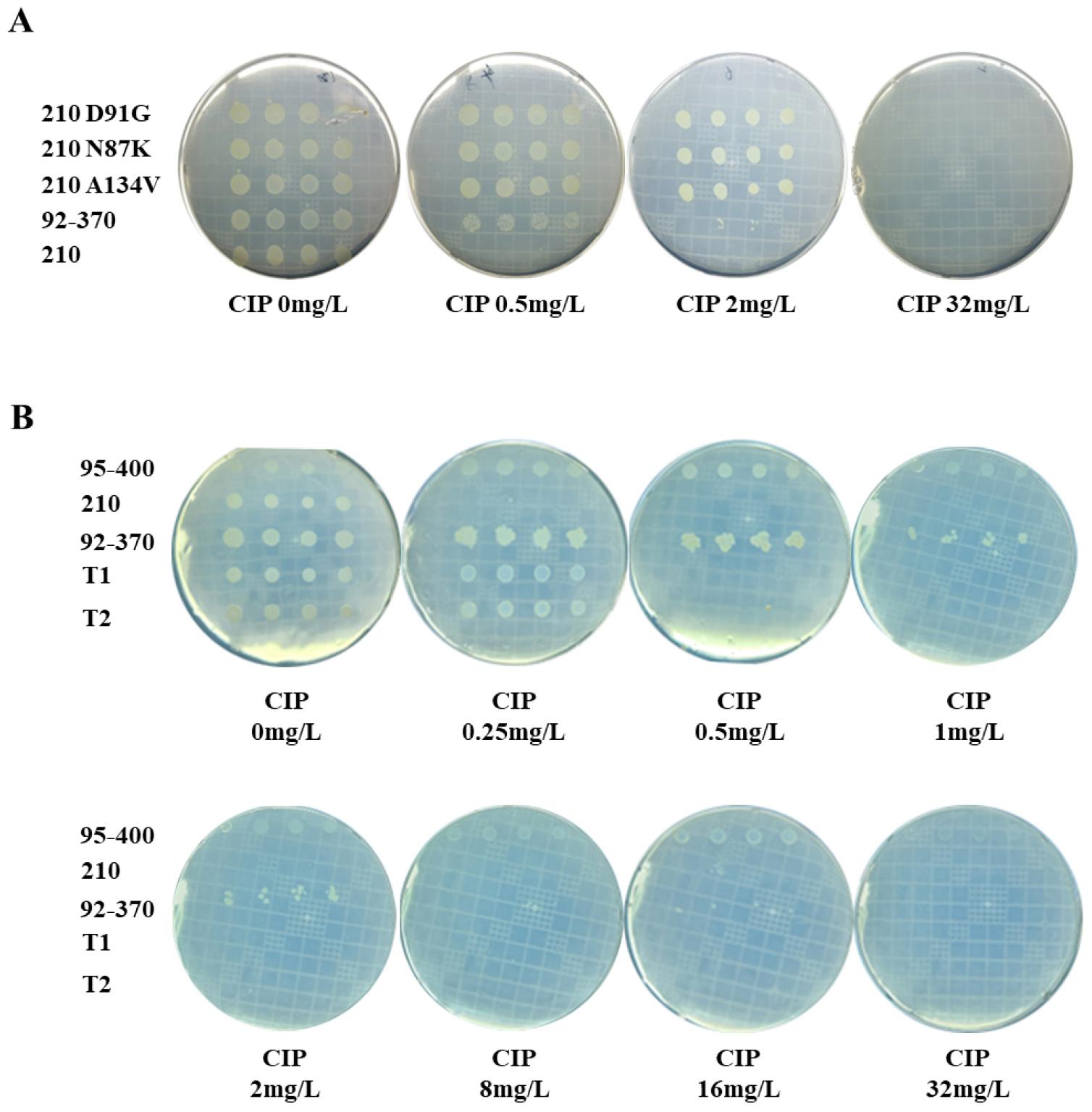
Fig. 4
MIC test for CIP of A134V mutation in strains 210. (A) MIC of A134V mutation obtained by natural transformation compared to single mutation strains D91G and N87K. DNA fragments containing the mutations of D91G, N87K, and A134V were subjected to natural transformation to quinolone-susceptible strains 210, and their MICs were confirmed using four transformants for each mutation. The WT strain 210 was used as a negative control, and the strain 92-370 containing the original A134V mutation was used as a positive control. (B). MIC test was conducted to validate the resistance level of the single-point mutation A134V.
From the aforementioned experiment, ten colonies of the wild type (WT) and nine colonies of A134V mutants were carefully selected, and their QRDR sequences were meticulously analyzed, as outlined in Table 3. Transformants exhibited mutations within the QRDR region, with a variety of mutations observed at 10 different residues, including D91G, D91N, D91A, D91Y, N87K, and G85D. Notably, strains resulting from the transformation of gyrA with A134V predominantly displayed mutations at position 91, with the D91G mutation being most prevalent in this context (Table 3).
Table 3.
List of mutations in QRDR of gyrA induced by natural transformation of gyrA with A134V mutation
| mutations | No. of strains | |
|---|---|---|
| naturally acquired gyrA mutation | D91G | 3 |
| D91N | 3 | |
| D91A | 1 | |
| D91Y | 1 | |
| N87K | 1 | |
| G85D | 1 | |
| mutation obtained by transformation of gyrA with A134V | D91G | 7 |
| N87K | 1 | |
| A88V | 1 |
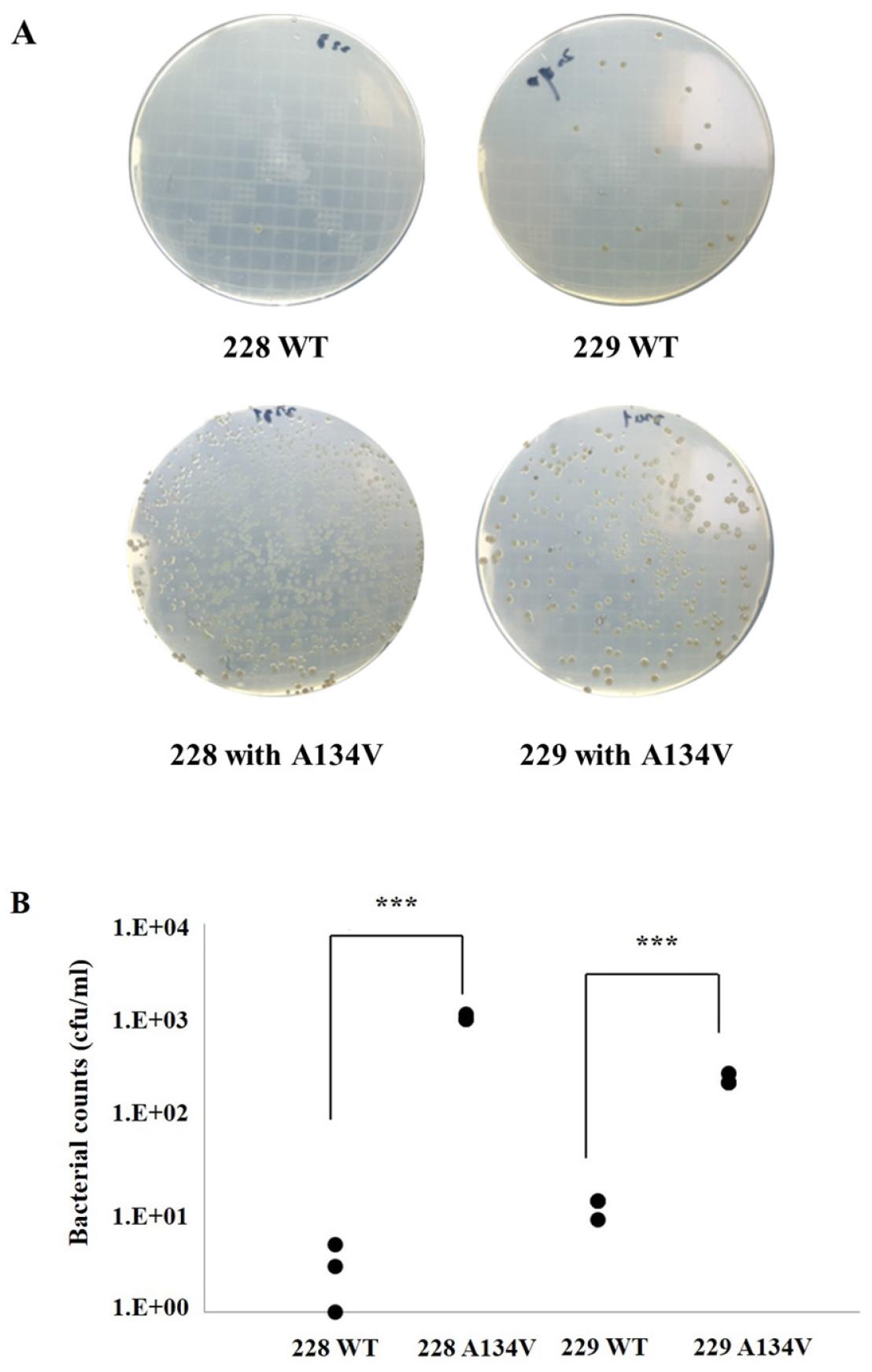
To validate whether the A134V mutation in gyrA induces mutations within the QRDR region during natural transformation, leading to susceptibility observed in strain 210, gyrA DNA fragments containing the A134V mutation were introduced into two other quinolone-susceptible strains, 228 and 229, akin to strain 210. Post-transformation, cells were cultured at a density of 4 × 10^5 colony-forming units (cfu) per plate on agar supplemented with CIP at 1 mg/L. For strain 228, the number of colonies with WT gyrA was determined to be 4.3 ± 1.4, whereas with the A134V mutant, it was notably higher at 1080 ± 301.4. Similarly, for strain 229 transformed with either WT gyrA or A134V mutants, 228 ± 63.6 and 12.3 ± 3.5 colonies grew on CIP-containing medium, respectively (refer to Supplementary Fig. 1 A, B for detailed results). These findings suggest that the A134V mutation in gyrA, inducing quinolone resistance, is not exclusive to susceptible strain 210 but also affects strains 228 and 229.
To mitigate potential confounding effects of spontaneous mutations within the QRDR of gyrA and assess the resistance conferred by the A134V mutant, a single mutation A134V DNA fragment, coupled with the kanamycin antibiotic resistance gene, was incorporated into the pGEM®-5Zf(+) vector for colony selection, obviating the use of kanamycin as a selection marker. Subsequently, the plasmid was introduced into susceptible strain 210. Two colonies harboring the A134V mutation (T1 and T2) were isolated, and the MIC of ciprofloxacin was determined. Concurrently, strains 95-400 and 210 served as negative controls, whereas strain 92-370 served as a positive control. The experiments were repeated thrice, and the results indicated that T1 and T2 exhibited low resistance to quinolone (MIC ~ 0.5 mg/L), similar to the susceptible strain 210, whereas the resistant strain 92-370 displayed a notably higher MIC of 8 mg/L (refer to Table 1 and Fig. 4B for detailed results).
Binding position of CIP with GyrA in present of mutations generated by molecular docking
The three-dimensional structures of GyrA protein variants with A134V, D91G, and N87K were individually predicted using the WISS-MODEL server (27), followed by molecular docking with CIP using the Autodock Vina program (28). The CIP binding pockets of the mutants are illustrated in Fig. 5B, C, and D, respectively. In the GyrA structure, the QRDR initiates with an alpha-helix and terminates with a beta-sheet structure. The position of residue 134 lies proximal to the beta-sheet structure of the QRDR and the CIP binding pocket (Fig. 5B). Upon superimposing the structure with the wild-type GyrA, the substitution of Alanine with Valine at position 134 induces movement in the side chain of L104, a constituent of the QRDR region. However, there is no discernible alteration in the remainder of the protein, particularly in the QRDR region and the CIP binding site.
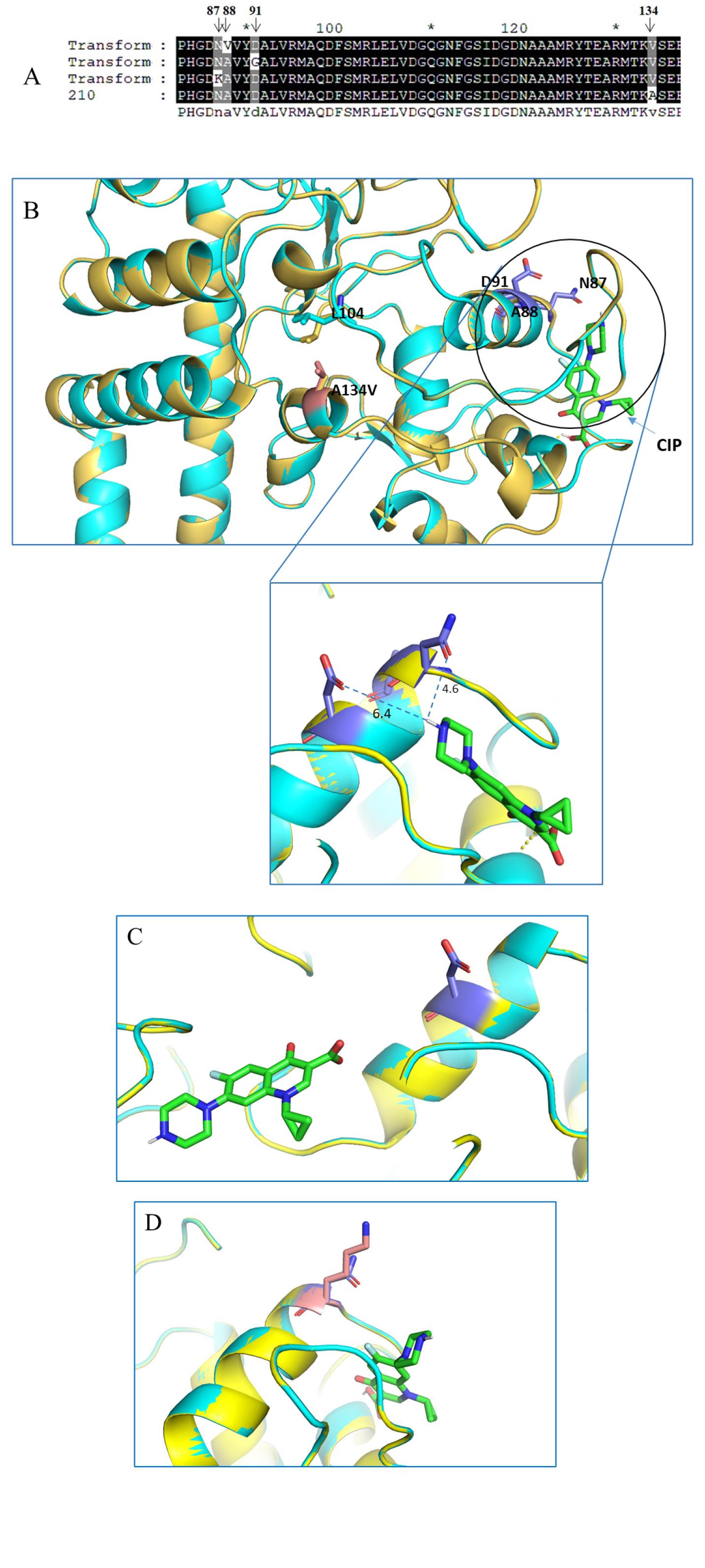
Fig. 5
Binding position of CIP with GyrA in present of mutations generated by molecular docking. (A) Sequence alignment of strains colonies selected randomly obtained from A134V mutagenesis by natural transformation. (B) A134 is located close to the GyrA active site. Point mutation of A134V disturbing the QRDR region determined by superposition of predicted structure of 210 GyrA with and without A134 mutation (shown in ribbon with yellow and cyan color, respectively). CIP binding location was determined by Autodock Vina. CIP molecule was shown in stick (green). QRDR point mutation residues N87, A88, and D91 were shown in purple. Mutated residue 134V was shown in salmon. (C) The binding position of CIP in present of D91G mutation predicted by molecular docking (D) The binding position of CIP in present of N87K mutation predicted by molecular docking.
The molecular docking of CIP with the wild-type protein structure reveals interactions between the carbonyl group of D91 and N87 and the amide group of CIP at distances of 6.4 Å and 4.6 Å, respectively. Simultaneously, the individual mutations of D91G and N87K in the GyrA protein structure were predicted separately. Fig. 5C and D demonstrate the differential positions of CIP binding with the GyrA protein in the presence of these QRDR mutations compared to the wild type. The increased distance and altered orientation of CIP result in a loss of binding affinity to residues within the QRDR region, especially in the cases of D91G and N87K mutants. This deviation in binding may potentially contribute to the reduced effectiveness of CIP against bacteria harboring these mutations, highlighting the significance of understanding the structural basis of antibiotic resistance.
DISCUSSION
DNA gyrase, a target of quinolones that hinders bacterial growth, participates in the negative supercoiling process during bacterial DNA synthesis (29). DNA gyrase comprises four subunits consisting of two gyrA and two gyrB units. Mutations at the QRDR position of either gyrA or gyrB lead to a reduced affinity for the enzyme-DNA complex, resulting in quinolone resistance (23, 29, 30). GyrA is generally recognized for its significant impact on quinolone resistance (16).
Consistent with previous studies, mutations at positions 87 (N87K) and 91 (D91G, D91N) in GyrA were prominent in quinolone-resistant strains, leading to significantly elevated minimum inhibitory concentrations (MICs) of LVX, CIP, and MOX over 8 mg/L, 8 mg/L, and 16 mg/L, respectively. These findings reinforce the critical role of QRDR mutations in quinolone resistance (16, 31, 32).
However, the moderate resistance observed in strain 92-370, in the absence of QRDR mutations, suggested alternative mechanisms of resistance (Table 1, Fig. 2). Instead, six candidate mutations (A134V, R190M, A705T, and N709S in gyrA and R249H and E624K in gyrB outside QRDR sequence have been found that can affect the quinolone resistance were identified in the strain 92-370. Interestingly, mutations outside the QRDR, particularly A134V in GyrA, emerged as a key finding. While A134V alone did not directly confer significant resistance, natural transformation experiments suggested that this mutation could facilitate the acquisition of spontaneous QRDR mutations during selective pressure from quinolones. This phenomenon highlights the potential of non-QRDR mutations to influence mutational landscapes, indirectly contributing to resistance. These results align with prior work suggesting that mutations beyond the QRDR can modulate quinolone susceptibility (23).
The three-dimensional structure of the GyrA protein with A134V, D91G and N87K mutations were independently generated by SWISS-MODEL (27), and the position of the CIP binding location was determined by Autodock program (28) (Fig. 5). In the GyrA 3D structure, QRDR starts with one alpha helix and ends with one beta-sheet structure. The position of residue 134 is in proximity to the beta-sheet structure position of the QRDR and the CIP binding pocket (Fig. 5). By structure superimposed with wild-type GyrA, the change from Alanine to Valine at position 134 caused the movement of the side chain of Leucine 104, which belonged to the QRDR region. Experimentally, none of the point mutations found in gyrA and gyrB were related to quinolone resistance, but the natural transformation of A134V mutation with the presence of CIP caused the spontaneous mutation to occur in the QRDR region (Fig. 5A) that led to a level of resistance increasing in susceptible strain 210. On the other hand, the molecular docking result showed the interaction between D91 and N87 of 6.4 and 4.6 Å, respectively by C=O group and NH group. The mutations of these two residues to neutral or alkyl group (refer to Table 3) led to reducing the binding affinity with CIP molecule and consequently, the drug could not bind to GyrA (Fig. 5C and 5D). Once again, this result suggested the role of point mutation of QRDR, particularly in this study, N87K, D91G dominantly appeared to be essential for quinolone resistance in H. pylori isolated from Korean pediatric patients.
Our findings also raise broader questions about the interplay between selective pressures and mutational hotspots. The increased frequency of colonies with QRDR mutations in strains transformed with A134V-mutant DNA suggests that selective environments may promote resistance evolution through complex pathways. These results underscore the importance of investigating the full sequence context of gyrA and gyrB in resistance studies, rather than focusing solely on QRDR sites In addition to genetic factors, non-mutational mechanisms, such as efflux pumps, may also contribute to quinolone resistance, as seen in other bacterial species like Pseudomonas aeruginosa, Escherichia coli, and Candida albicans (33). Recently, a molecular biological diagnosis method for the QRDR site of gyrA has been proposed to identify quinolone-resistant H. pylori, and our results show that such a method needs to be expanded to a wide area, including the QRDR part. This can be an important basis for the research focused on the limited mutation of the QRDR site in the situation where the application of quinolone to the treatment of H. pylori increases.
CONCLUSION
Our study confirmed that mutations in the non-QRDR of gyrA and gyrB genes in H. pylori are not directly linked to quinolone resistance. This finding emphatically underscores the crucial role of the QRDR in mediating resistance to quinolone antibiotics. Notably, our analysis of strain 92-370, which displayed moderate resistance to ciprofloxacin without QRDR mutations, supports this conclusion and suggests the possibility of resistance being influenced by other factors. Furthermore, the study reveals that specific mutations outside the QRDR, such as the A134V mutation in gyrA, can impact the mutation rate within the QRDR. This indicates a complex interplay where non-QRDR mutations, especially those near the QRDR, may indirectly contribute to increasing resistance levels by facilitating spontaneous mutations within the QRDR. These findings not only contribute to a better understanding of antibiotic resistance mechanisms in H. pylori but also highlight the need for comprehensive genomic analyses in developing effective treatment strategies. Our research paves the way for future investigations into the intricate genetic factors influencing antibiotic resistance and guides clinical management of H. pylori infections, emphasizing the necessity of targeting both QRDR and adjacent regions in diagnostic and therapeutic approaches.