INTRODUCTION
One of the complications that may develop in ventilated patients is ventilator‐associated pneumonia (VAP), which contributes to approximately half of all cases of hospital-associated pneumonia and is the second most common nosocomial infection in the intensive care unit (ICU) (1). Several different routes of infection by VAP pathogens have been proposed, but in most cases infection appears to result from aspiration of the oropharyngeal microflora or colonizing pathogens (2). Intubation and critical illness reduce oral immunity and can exacerbate poor oral hygiene (3, 4). Dental plaque accumulates rapidly in the mouths of critically ill patients and colonization by microbial pathogens is likely as the amount of plaque increases (5, 6). Microaspiration from oropharyngeal microbes are thought to play a role in the etiology of VAP (7) and the high prevalence of oral colonization with VAP-associated pathogens have been reported (8). VAP is often a clinical diagnosis without confirmation by bacterial culturing and subsequent identification of putative pathogens (9). In a study of mechanically ventilated patients, 80% of culture-negative respiratory samples had high abundances of oral bacteria (10). Several studies have shown that maintenance of oral hygiene in patients under mechanical ventilator can reduce the rate of VAP (11, 12). Thus, understanding the oral microbiome dynamics in intubated patients should help prevent or manage VAP in ICU patients.
16S ribosomal RNA (rRNA) gene sequencing has allowed culture-independent characterization of the bacterial communities in health and disease (13, 14, 15). Although oral microbiome has been extensively studied in oral diseases such as periodontitis and caries, it has not been extensively studied in intubated subjects under mechanical ventilator. Here, we performed serial 16S rRNA gene sequencing of buccal samples from intubated patients under mechanical ventilator. Our goals were to identify features of bacterial community structure in intubated subjects and characterize the dynamics of the oral bacterial microbiome during mechanical ventilation.
MATERIAL AND METHODS
Participants
The inclusion criteria for ICU patients were; 1) > 18 years of age, 2) endotracheal tube use with holding methods, 3) consent for participation from the patient’s family. For healthy control, age matched 13 volunteers (7 male, 6 female, age 67.5 ± 7.4) without any systemic diseases were recruited. The enrolled intubated patients (9 male, 1 female, age 68.8 ± 11.2) include a heterogeneous set of underlying diseases (Table 1). Average observation period was 10.7 ± 12.4 days (range: 1-41 days) and 7.9 ± 6.9 (range :1-21) samples per patients were collected. Broad spectrum antibiotics were administered to the patients. Carbapenem (7/10) which include ertapenem and meropenem was most frequently prescribed antibiotics followed by penicillin (6/10) including piperacillin-tazobactam (Table 1). For longitudinal microbiome analysis, several timepoints were selected as follows; initial samples collected within 2 days (Day2), 1st week samples (1 wk), 2nd week samples (2 wk), 3rd week samples (3 wk) and samples collected over 3 weeks (over 3 wk).
Table 1.
Characterization of subjects under mechanical ventilation
This study was approved by the Institutional Review Board (DAUHIRB-19-072). Intubated participants in this study were mostly sedated or at severely poor levels of consciousness; therefore, family members were considered as the legal representatives with the power of attorney based on the International Council for Harmonization (16). During the participant recruitment period, the purpose, voluntary nature of participation, confidentiality of information, and procedures used in the study were explained to family members, and informed consent was obtained from each family.
Oral swab sample preparation
Buccal swap samples were collected using the Levine technique; the swab was rotated over an approximately 1cm2 area with sufficient pressure (17). Sterile cotton swabs were used for the collection and were placed in a 15 mL conical tube labelled with the sample ID and stored at –80°C.
Extraction of Genomic DNA and Next Generation Sequencing
Samples were suspended in phosphate buffered saline (PBS), vortexed, and then centrifuged. DNA was extracted using a Gram positive DNA purification kit (Lucigen, Biosearch Technology, Novato, CA) following the manufacturer’s instructions. The final concentration was measured with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA) and stored at −80°C until use. PCR amplification of the 16S ribosomal RNA gene V3-V4 region (341F-785R) was performed. The primer sequences used for amplifications were as follows: 341F: 5’-CCT ACG GGN GGC WGC AG-3’, 785R: 5’-GAC TAC HVG GGT ATC TAA TCC-3’(18). Purified amplicons were pooled in equimolar and paired-end sequenced with MiSeq (Illumina, San Diago, CA, USA).
Bioinformatic Analysis, Statistical Analysis, and Visualization
Basic microbiome analyses have been performed using the QIIME2 (version 2020.6) (19) and associated plugins. To measure alpha diversities, Faith PD index, Choa1 index, and Shannon’s index method were used. For the beta diversity analysis, the weighted UniFrac principle coordinates analysis (PCoA) was used. The Kruskal-Wallis test was used to assess the statistical significances between the groups for alpha diversity and relative abundance. To assign taxonomy to the unique representative sequences, pre-trained Naive Bayes classifier, using Human Oral Microbiome Database (eHOMD) 16S rRNA Extended RefSeq sequences (version 15.1) (20), were used. To test differential abundance of bacterial species among test groups, linear discriminant analysis effect size (LEfSe) (21) was applied with default settings.
RESULTS
Diversity and abundance of microbiota
The alpha diversity of the microbiota was estimated by Chao1 (Fig. 1A), Faith PD (Fig. 1B), and Shannon test (Fig. 1C). In intubated patients under mechanical ventilator, richness (Chao1 and Faith PD) of the microbiota was significantly reduced compared to the healthy subjects (Fig. 1A and 1B). The evenness (Shannon) of intubated patients were reduced compared to the healthy subjects but statistical significance was not prominent (Fig. 1C). To visualize the overall microbiome structure, weighted UniFrac PCoA was applied. The microbiome composition was significantly different between healthy subjects and intubated patients (Fig. 1D).
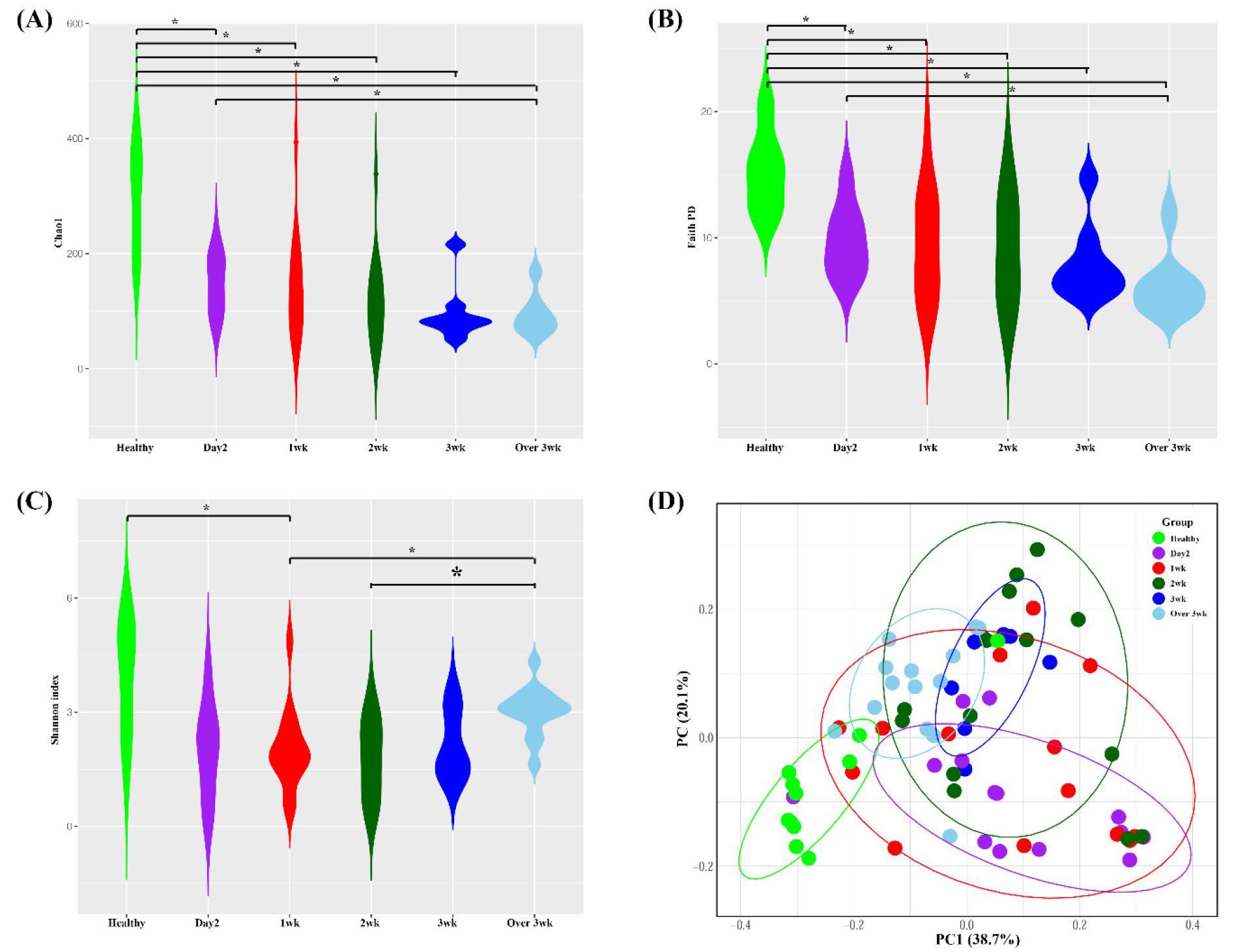
Fig. 1
Bacterial community comparisons. Buccal samples were collected from healthy and intubated patients. (A) Chao1 alpha diversity (B) Faith’s PD alpha diversity (C) Shannon alpha diversity, (D) Weighted UniFrac Principle Coordinate beta diversity. *p < 0.05
Next, abundance of the microbiota was analyzed. At the phyla level, the five most abundant were Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria, which represented more than 99% of the total sequences. The abundance of Actinobacteria was most abundant in intubated patients within 2 days and their abundance showed gradual decrease. Compared to healthy subjects, the abundance of Bacteroidetes and Firmicutes was decreased in intubated patients during the treatment (Fig. 2A). At the genus level, the most abundant genera in the healthy subjects were Streptococcus, Gemella, Neissseria, Heamophilus, and Klebsiella, whichconstituted more than 70% of the total sequences. In intubated patients, Corynebacterium,Acinetobacter, Streptococcus, Staphylococcus, and Enterococcus represented more than 70% of the total sequences. In intubated patients, the abundance of Corynebacterium and Enterococcus was most abundant in day2 group and showed gradual decrease while the abundance of Acinetobacter and Staphylococcus showed gradual increase from initial treatment to week 3 (Fig. 2B).
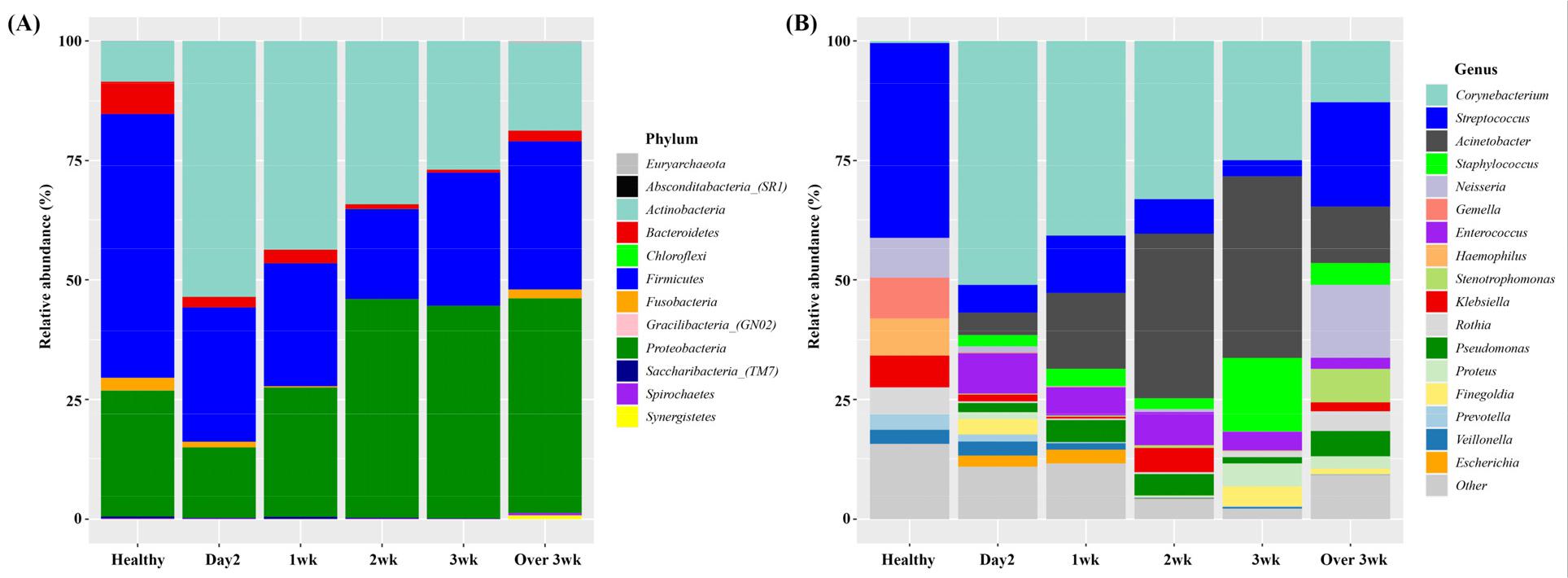
Fig. 2
Relative abundance of bacterial species in healthy and intubated patients at various time points. (A) Phylum level and (B) Genus level. Day2: samples collected within initial 2 days intubated, 1 wk: samples collected within first week, 2 wk : samples collected within second week, 3 wk : samples collected within third week, over 3 wk : samples collected over 3 week.
Species taxa comparison
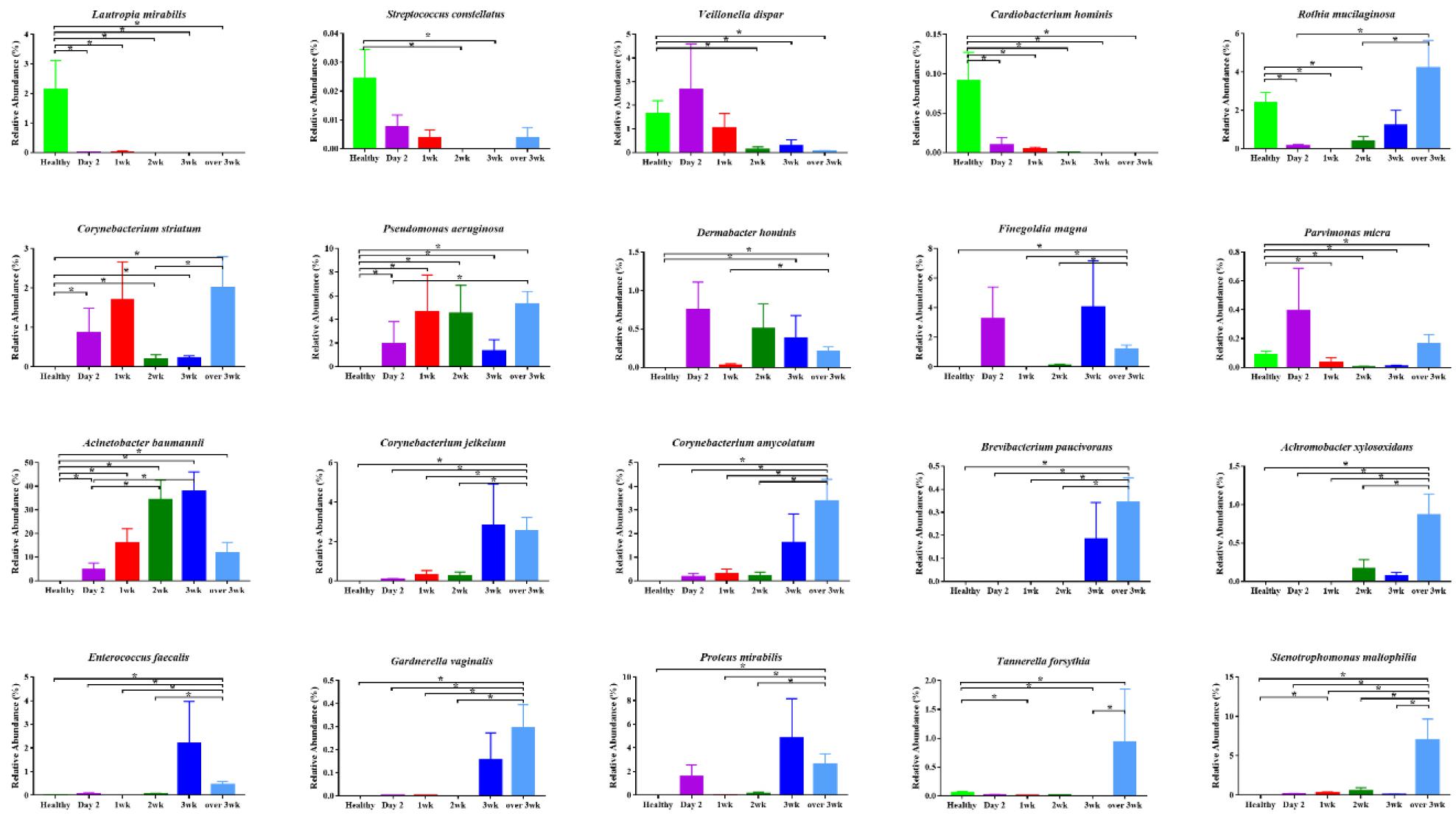
LEfSe was applied to evaluate the differences in bacterial abundance during ventilator treatment in incubated patients. In initial day2 samples, the abundance of Corynebacterium, Veillonella dispar, Prevotella, and Campylobacter gracilis were significantly higher compared to other time-points. In 1st week samples, the abundance of Campylobacter, Actinomyces, and Capnocytophaga were significantly higher compared to other time-points. In 3rd week samples, the abundance of Acinetobacter baumannii, Staphylococcus, Proteus mirabilis, and Enterococcus faecalis were significantly higher compared to other time-points. In over 3 week samples, the abundance of Pseudomonas aeruginosa, Corynebacterium amycolatum, Tannerella forsythia, and Gardnerella vaginalis were significantly higher compared to other time-points (Fig. 3A and 3B). Finally, the abundance of significant species identified by LEfSe was validated. The abundance of Lautropia mirabilis, Streptococcusconstellatus, and Cardiobacterium hominis was abundant in healthy subjects and showed abrupt decreased following intubation. The abundance of Acinetobacter baumannii showed gradual increase from day2 to week 3 in intubated patients. Several bacteria showed significant increase in samples over 2 weeks (week 3 and over 3 week samples), e.g. Corynebacterium jeikeium, C. amycolatum, Brevibacterium paucivarans, Achromobacterxylosoxidans, Enterococcus faecalis, Gardnerella vaginalis, Proteus mirabilis, Tannerella forsythia, and Stenotrophomonas maltophila (Fig. 4).
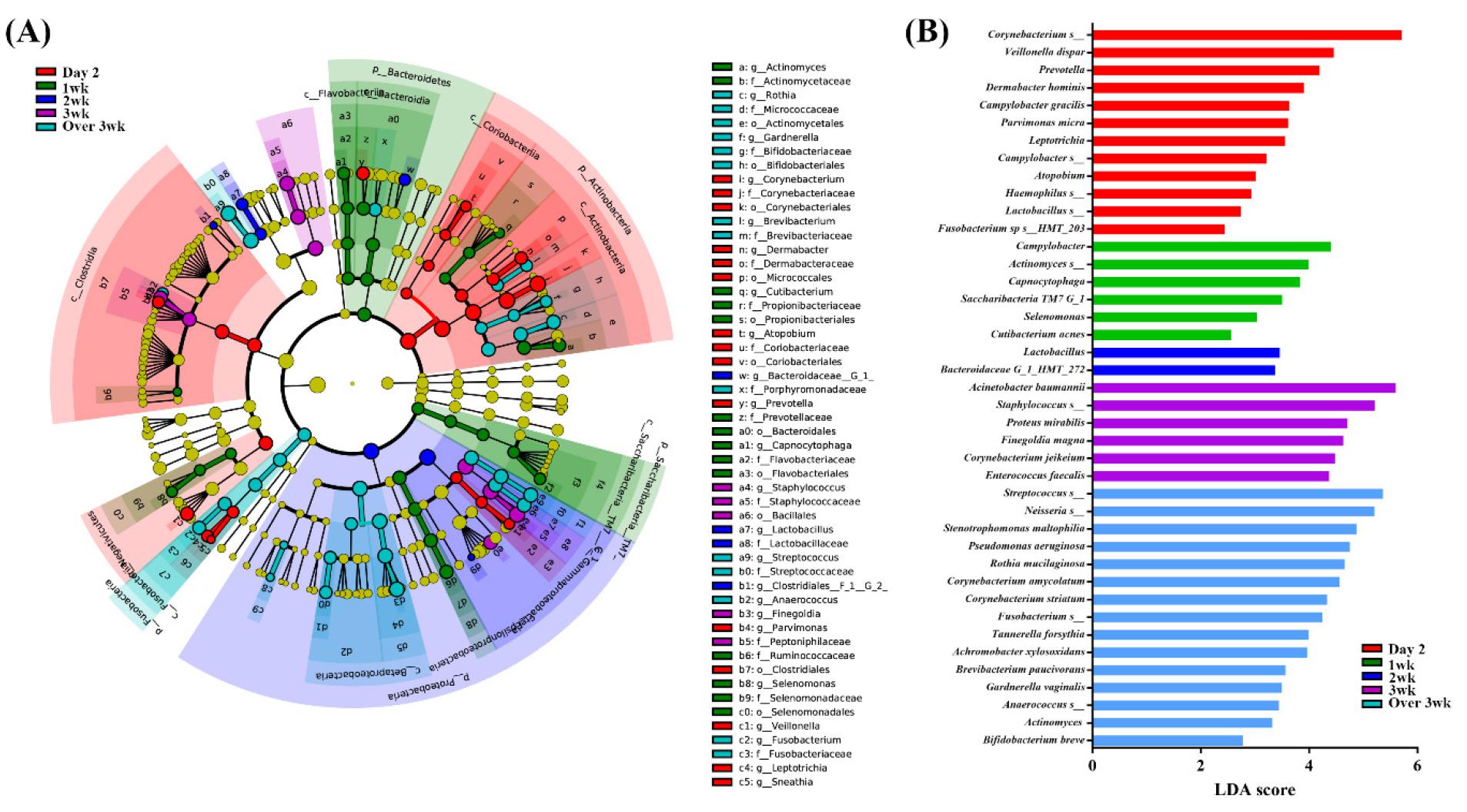
Fig. 3
Comparisons of microbiota depending on intubation period that presented significantly different contents. (A) Cladogram of significant species. (B) Linear discriminant analysis (LDA) score of significant species. The analysis has been performed using linear discriminant analysis effect size (LefSe) method. Day2: samples collected within initial 2 days intubated, 1 wk: samples collected within first week, 2 wk : samples collected within second week, 3 wk : samples collected within third week, over 3 wk : samples collected over 3 week.
DISCUSSION
Intubation and the presence of the endotracheal tube can exacerbate poor oral hygiene in ICU patients receiving mechanical ventilation treatment (3, 4). Pulmonary complications of mechanical ventilation include VAP (22). Respiratory pathogens can colonize the oral cavity prior to entering the lower respiratory tract (23). For efficient VAP prevention and management, it is imperative to understand the features of bacterial community structure and the dynamics of the oral bacterial microbiome during mechanical ventilation. In this study, we analyzed longitudinal oral microbiome change to understand the microbiome dynamics in intubated patients.
To characterize the initial oral microbiome change in intubated patients, healthy participants were recruited for comparison. Since oral microbiome dynamically change during mechanical ventilation, several time points were selected for the analysis. Compared to healthy subjects, the richness (Chao1 and Faith PD) and evenness (Shannon index) of the microbiota was significantly decreased in intubated patients. A course of antibiotics can lead to a marked reduction in species diversity and expansion of relative abundance of certain taxa (24). Since most of the patients enrolled in this study had been on antibiotic therapy, effect of antibiotics could have influenced the oral microbiome diversity. Also when beta-diversity was determined, the oral microbial structure was significantly different between healthy subjects and intubated patients suggesting the oral microbiome structure was significantly distorted in intubated patients.
When overall abundance was determined, Corynebacterium,Acinetobacter, Streptococcus, Staphylococcus, and Enterococcus represented more than 70% of the total sequences in samples from intubated patients. The abundance of Corynebacterium was most abundant in Day2 samples and remained highly abundant in intubated patients during the observation. The relative abundance of the genus Corynebacterium have been reported to be significantly higher in the pneumonia compared to healthy subjects (25). At the species level, the overall microbiome structure of intubated patients was significantly different from healthy subjects. When LEfSe was applied to compare significant taxa between healthy and intubated patients, a very long list of bacteria taxa was produced. Thus, we focused significant taxa within intubated patients. At each time point, there were several bacteria taxa significantly increased compared to other time points. The abundance of Veillonella dispar, Campylobacter gracilis, Acinetobacter baumannii, Proteus mirabilis, Enterococcus faecalis Pseudomonas aeruginosa, Corynebacterium amycolatum, Tannerella forsythia, and Gardnerella vaginalis were among the significant taxa in intubated patients. A. baumannii is an opportunistic human pathogen that predominantly infects critically ill patients, which causes a range of nosocomial infections across multiple anatomical sites and most commonly it manifest as ventilator-associated pneumonia or central line-associated blood stream infections (26). Especially, Acinetobacter have been noted to be a primary pathogen of VAP in some Asian countries (27, 28).
During longitudinal observation, pathogenic bacteria including A. baumanni, Enterococcus, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus were also frequently found. In a meta-study that included 24 investigations conducted with ventilated patients, gram negative bacteria represented 58% of recovered organisms in VAP patients. The predominant gram negative bacteria were P. aeruginosa and Acinetobacter spp., followed by Proteus spp., Escherichia coli, Klebsiella spp., and H. influenzae. Among gram positive bacteria, S. aureus was involved in 20% of the cases (22). Moreover, known VAP pathogens such as H. influenzae, Escherichia sp.,S. pneumoniae, Proteus mirabilis, and Pseudomonas sp. have been identified in the oral cavity from patients with VAP (23). Thus, strong coincidence of pathogens in oral cavity and VAP supports the need to monitor oral microbiome to prevent or manage VAP in intubated patients.
When the microbiome change of the significant taxa was validated, oral microbiome commonly found in healthy subjects showed abrupt decrease in intubated patients. The abundance of A. baumannii showed gradual increase in intubated patients and there were some bacteria that showed significant increase in samples over 2 weeks (week 3 and over 3 week samples). Thus, there is continuous oral microbiome change in intubated patients. There are several limitations to this study design. First, the study uses a relatively small sample size. A limited number of observable patients were available for participation, because intubated patients usually underwent a tracheostomy within 2 weeks, and even more critical patients were directly selected for tracheostomy. Our findings should be validated in a larger clinical cohort. Although power calculations were not feasible in this study, an adequate number of participants would be needed to allow stratification of participants into different predisposing medical conditions or antibiotics. Second, a well-designed longitudinal study with adequate sample size is needed to elucidate the role of oral microbiome in VAP development or clinical outcome.
The present study provides valuable information concerning the oral microbiome in intubated patients. A better understanding of the sequential microbiome change in oral cavity of intubated patients and the role of oral bacteria in VAP development should provide better understanding for effective prophylactic and therapeutic interventions for VAP.