INTRODUCTION
The gut microbiome, a vast and diverse microbial ecosystem has captured significant interest for its profound impact on human health. Recent epidemiological, physiological, and animal studies over the past decades have demonstrated that the gut microbiome is a significant determinant in nearly all human diseases and phenotypes, much like our own genes (1, 2, 3). Despite this critical role, progress in elucidating the mechanistic pathways and developing microbiome-based therapies has not advanced as expected (4).
Current research on gut microbiome predominantly relies on culture-independent metagenomic next-generation sequencing (mNGS) methods (5). Metagenomics studies are essential for understanding gut microbiota diversity (6). However, metagenomics also highlights that a large proportion of gut microbes remains uncultured (7, 8, 9, 10). This gap necessitates complementary culture-dependent experiments to fully understand microbial functions (11, 12, 13) (Fig. 1). Culturing specific gut microbes identified through metagenomics is vital to determine their causative roles in the diseases and phenotypes of human. This approach aligns with Koch’s postulates (14, 15). Moreover, the functions of gut microbes cannot be fully understood without pure cultures of the implicated species. (16). Pure cultures are the first step for whole-genome sequencing (WGS), which provides valuable reference genomes (17). These references improve the resolution of metagenomic analyses, yet the lack of complete genomes from cultured species remains a significant limitation (18).
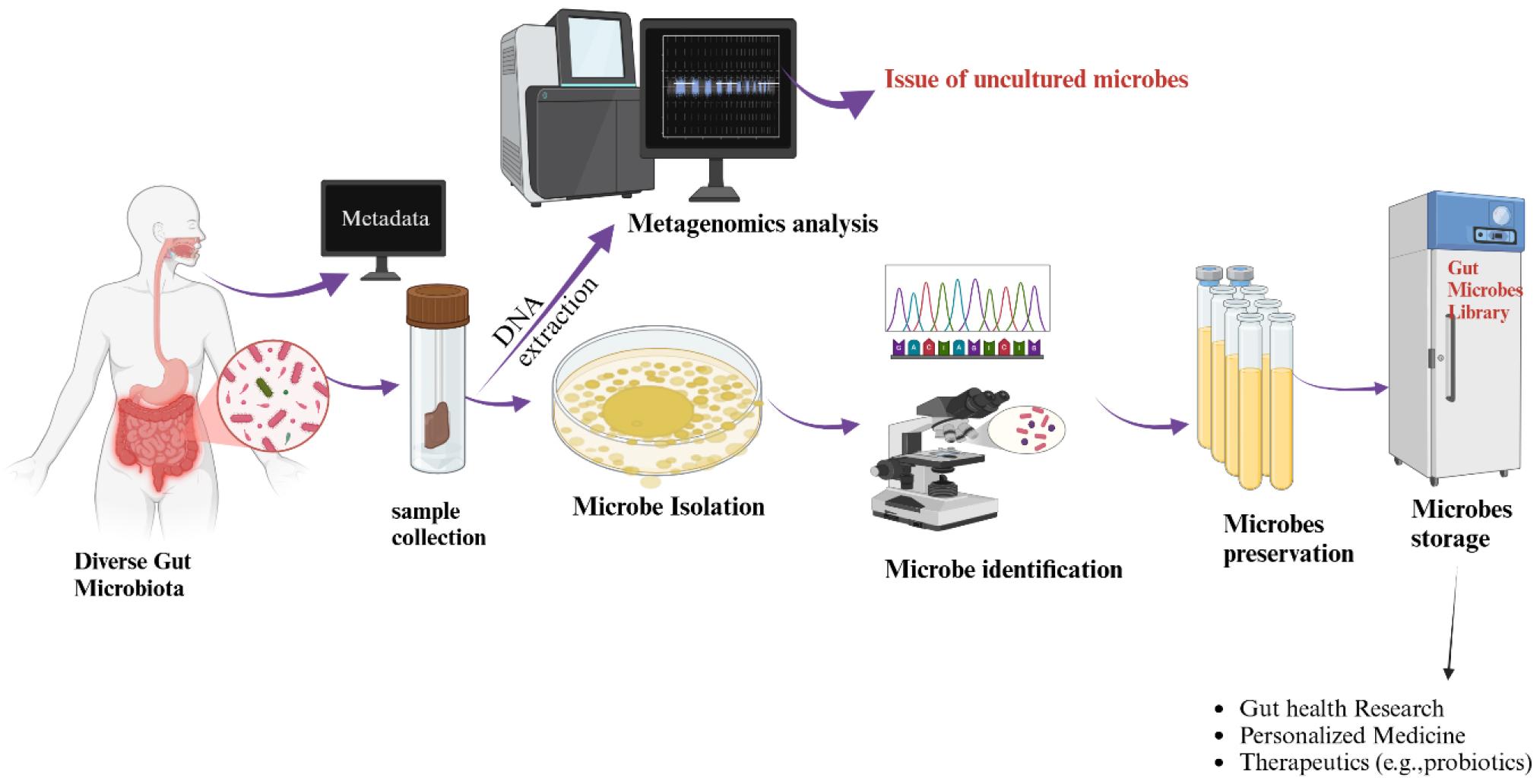
Fig. 1
Conceptual workflow depicting the need for a gut microbe library to overcome the limitations of metagenomic sequencing.
Despite the urgent need for gut microbe libraries to elucidate gut microbiome functions, establishing a comprehensive gut microbe library that covers a significant portion of the human gut microbiome remains highly challenging. This is because most gut microbes in the human gut microbiome do not grow in existing culture media and remain unculturable. Due to the unculturable nature of many gut microbes, various complex culture media, including Brain Heart Infusion (BHI), Gut Microbiota Medium (GMM), Fastidious Anaerobe Broth (FAB), Bacterial Growth Medium (BGM), and Yeast Casitone Fatty Acid (YCFA), have been developed to support their growth. However, these media are far from universally supporting the growth of all gut microbes. Beyond culture media limitations, the requirement for strict anaerobic conditions and the challenges of long-term storage further complicate the establishment of a comprehensive gut microbe library.
Reflecting current trends in gut microbiome research, we discuss the significance of establishing a comprehensive gut microbe library as an essential microbial resource for elucidating human gut microbiome functions. It discusses existing gut microbial libraries and outlines the key components necessary for their development. Additionally, the perspective explores the potential applications of gut microbe culture collections in areas such as disease modeling, microbiome-based therapeutics, personalized medicine, and advancing our understanding of host-microbe interactions. Moreover, it examines the potential of artificial intelligence (AI) to accelerate microbiome research advancements.
THE SIGNIFICANCE OF ESTABLISHING A COMPREHENSIVE GUT MICROBE LIBRARY AND CURRENT CULTURE COLLECTIONS OF GUT MICROBES
The limitations of culture-independent research in elucidating the functions of the human gut microbiome, which relies primarily on metagenome sequencing, are driving a new era of proof-of-concept research where a comprehensive gut microbe library is a prerequisite. Pure cultures of gut microbes identified through culture-independent methods provide an essential starting point for determining whether these organisms meet Koch’s postulates as causative agents of specific diseases or phenotypes (19). Establishing a cause-and-effect relationship for specific gut microbes based on Koch’s postulates provides definitive evidence of the human gut microbiome’s function. Moreover, obtaining a pure culture of a specific gut microbe is the first step toward whole-genome sequencing, which is valuable in its own right and also serves as a reference sequence for mNGS. The genome sequence data from whole-genome sequencing would dramatically enhance the resolution of gut microbiome analyses to elucidate the function of the human gut microbiome in culture-independent analyses (20).
Beyond serving as a critical tool for elucidating the human gut microbiome, the gut microbes in a comprehensive gut microbe library can be utilized as drug substances in microbiome-based therapeutics. The culture collections of a gut microbe library, accompanied by complete donor metadata, could potentially replace fecal microbiota transplantation (FMT), which remains a blunt tool for gut microbiome engineering. Thorough screening of donor gut microbiomes for pathogenic species is essential to ensure FMT safety, especially after the detection of SARS-CoV-2 in fecal samples during the COVID-19 pandemic (21). However, with a comprehensive gut microbial library, specific beneficial microbes could be selectively identified and used, enabling targeted and precise microbiome modulation for improved safety and efficacy. For instance, a recent study demonstrated that a combination of bacterial species, specifically Oribacterium sp. GMB0313 and Ruminococcus sp. GMB0270, provided complete protection against SARS-CoV-2 infection by stimulating CD8+ T cell-mediated immunity (22). Another recent study demonstrated that a consortium of 18 commensal bacterial strains (F18 mix) effectively suppressed the growth of antibiotics resistant Klebsiella Pneumoniae and other Enterobacteriaceae in germ-free mice by depleting essential nutrients required for the pathogen’s survival (23). This underscores the potential of developing a comprehensive gut microbial library, which would enable the selection of specific microbial strains with targeted functions—such as nutrient competition—to prevent or control pathogenic infections more precisely.
Although the significance of establishing a comprehensive gut microbe library cannot be overstated, the lack of appropriate culture media, the highly specific growth conditions required, and the strict long-term storage needs of gut microbes continue to hinder its development. Despite these challenges, several comprehensive gut microbial libraries have been established due to the importance of proof-of-concept research and the need for drug substances in microbiome-based therapeutics. For example, the Unified Human Gastrointestinal Genome (UHGG) collection comprises 204,938 non-redundant genomes from 4,644 prokaryotic species (24), while the Human Reference Gut Microbiome includes 5,414 distinct species (25). However, over 70% of UHGG species lack cultured representatives, limiting our understanding of gut microbe functions and host interactions. Recent large-scale cultivation efforts have successfully cultured 1,170 strains representing 400 human gut microbial species (26). These libraries provide extensive genomic and functional datasets that support comparative genomics, functional annotation, and evolutionary studies of gut microbiota. However, despite their broad scope, these resources face limitations, including incomplete representation of microbial diversity, underrepresentation of uncultivable or low-abundance taxa, and a notable lack of information on fungal species within the gut microbiome.
Although these gut microbe libraries have collected a substantial number of microbial strains, none can be classified as a truly comprehensive gut microbe library, except for GM Bank, which has collected 2,199 strains (781 bacterial species and 37 fungal species) (www.gmbank.org) (27). Table 1 provides an overview of the key features of currently available gut microbe libraries worldwide.
Table 1.
Gut Microbial Reference Libraries with descriptions, limitations, and reference
| library | Description | Limitations | Reference |
|---|---|---|---|
| GM Bank | Comprehensive gut microbe library with cultured 781 bacterial species and 37 fungal species | It is based on 16S rRNA and ITS sequencing, not whole genome sequencing, and lacks metadata | (27) |
|
Unified Human Gastrointestinal Genome (UHGG) Collection | A comprehensive catalog of 204,938 reference genomes from the human gut microbiome. | Limited to only rough genome information. Lack of cultural collection | (28) |
| Human Microbiome Project (HMP) | Provides a wide array of genomic and functional data on human-associated microorganisms. | Limited annotated species and functions | (29) |
| Culturable Genome Reference (CGR) | Includes 1,520 reference genomes from cultivated human gut bacteria asian metagenomes, enabling functional studies and deeper insights into gut microbiota. | Limited to cultivated bacterial strains, potentially missing uncultivable or rare microbes present in the gut microbiome. | (24) |
| Culturable Genome Reference 2 (CGR2) | An expanded version of CGR, providing 3,324 high-quality draft genomes from isolates. | Despite expansion, certain fastidious or rare microbes may remain uncultured | (20) |
|
Human Gut Microbial Biobank (hGMB) | Contains 1,170 strains representing 400 human gut microbial species. | Limited by the cultivability of certain gut microbes and potential underrepresentation of rare species. | (26) |
|
Human Gastrointestinal Bacteria Culture Collection (HBC) | A comprehensive set of 737 whole-genome-sequenced bacterial isolates from the human gut microbiome, providing a valuable resource for microbiome researchers. | miss uncultured or rare species and lack fungal species. | (30) |
|
Culture Collection of Human Gut Bacteria by the Wellcome Sanger Institute | A collection of gut bacterial isolates cultured from fecal samples, with accession numbers deposited in public culture collections, facilitating research and comparative studies. | Limited to strains that can be cultured from fecal samples, possibly omitting bacteria that are difficult to culture or present in low abundance. | (31) |
|
Extensive Personal Human Gut Microbiota Culture Collections | Utilizes high-throughput anaerobic culturing techniques combined with metagenomics to capture a significant proportion of an individual’s fecal microbiota, aiding personalized microbiome studies | While capturing a substantial fraction, it may still miss certain anaerobic bacteria or those requiring specific growth conditions not replicated in the study. | (32) |
|
A Library of Human Gut Bacterial Isolates Paired with Longitudinal Multiomics Data | Enables mechanistic microbiome research through a combination of cultured isolates and longitudinal data. | Limited by the diversity of isolates and the geographic representation of sample populations. | (33) |
CHALLENGES IN ESTABLISHING A GUT MICROBE LIBRARY
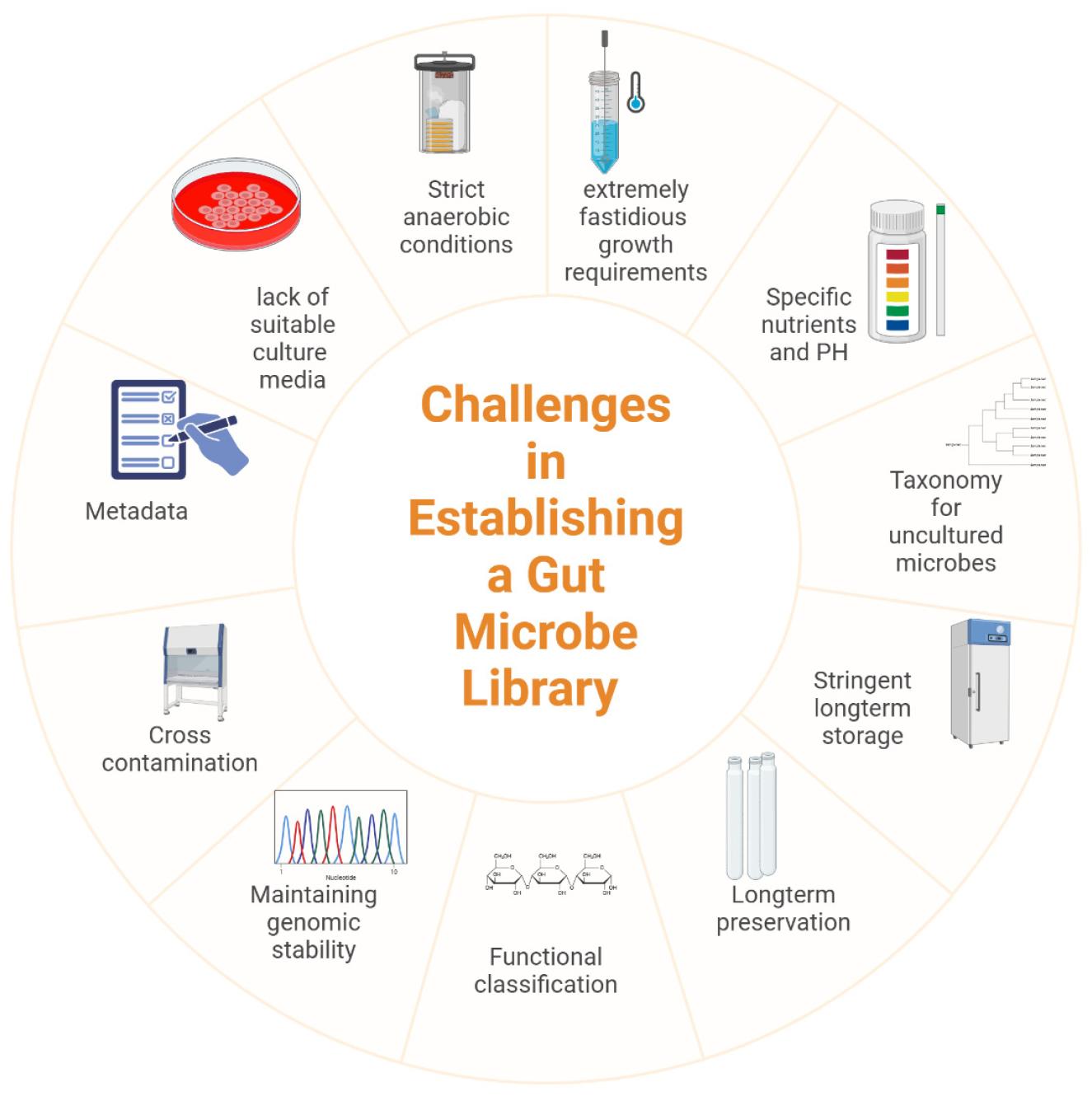
Establishing a comprehensive gut microbe library presents several challenges, including the lack of suitable culture media, extremely fastidious growth requirements, and stringent long-term storage conditions of gut microbes (34). Furthermore, the difficulty of culturing many gut microbes due to their strict anaerobic requirements. These microbes often need highly specific nutrients and pH levels. For instance, certain beneficial bacteria flourish in a colonic pH range of 5.5 to 7, and deviations can inhibit their growth (35, 36). Additionally, stringent long-term storage conditions, such as maintaining low temperatures and anaerobic environments, are necessary to preserve microbial viability and genetic stability over time (37). Preservation for long-term storage remains a significant challenge. Because gut microbes are highly sensitive to environmental changes, which can affect their viability and genomic integrity (38). There is also a notable lack of universal preservation media suitable for maintaining a wide range of gut microbes (39).
High costs, labor-intensive processes, and the need for specialized equipment further complicate efforts (40). Additionally, ensuring accurate taxonomic classification, functional annotation, and maintaining genomic stability over time are critical hurdles. These challenges restrict the cultivation and study of numerous uncultured species, limiting our understanding of microbial diversity and function (Fig. 2).
ARTIFICIAL INTELLIGENCE IN THE CONSTRUCTION OF GUT MICROBIAL LIBRARIES
Artificial intelligence (AI) and machine learning (ML) have emerged as powerful tools in microbiome research (27, 41). AI and ML revolutionizing data analysis, predictive modeling, and functional characterization (42). AI-driven bioinformatics pipelines can analyze vast metagenomic datasets, classify microbial taxa, and identify novel microbial strains with enhanced precision (43). Deep learning algorithms facilitate the functional annotation of genes and metabolic pathways, improving our understanding of microbial roles in health and disease (44).
A primary application of AI and ML in microbiome research is the development and utilization of comprehensive gut microbial libraries. These libraries serve as foundational datasets for understanding microbial diversity and function. AI models analyze sequencing data to discover new microbial taxa, antimicrobial resistance profiling, predict functional gene annotations, and reconstruct microbial genomes with high accuracy (44, 45, 46). Tools like QIIME2 (47) and DeepMicrobes (48) utilize advanced algorithms for taxonomic classification, functional annotation, and microbiome profiling. These tools contribute significantly to the construction and refinement of comprehensive gut microbial libraries.
AI also accelerates the curation of gut microbial libraries by integrating multi-omics datasets, such as metagenomics, metatranscriptomics, and metabolomics. For example, the UHGG collection benefited from machine learning algorithms that helped classify and annotate thousands of microbial genomes from human gut samples (28). Despite these advancements in Artificial intelligence (AI) and machine learning (ML) constructing comprehensive gut microbial libraries remains a challenge. These challenges include biases in training datasets, metadata, computational demands, and the need for standardized frameworks to ensure accurate and consistent genome curation.
CONCLUSION
The construction of a comprehensive gut microbes library is essential for advancing microbiome research. It will enable a detailed understanding of gut microbial composition, diversity, and function. Despite advances in sequencing, multi-omics technologies, and culture-based methods, cultured gut microbes remain a challenge. Standardized protocols for sample collection, storage, culturing, and genomic characterization are crucial for building a reliable microbial reference database.
Future efforts should prioritize developing versatile culture media that closely mimic the gut environment. Synthetic biology and metabolomics can help identify essential growth factors, while co-cultivation and microfluidic technologies may facilitate the isolation of previously unculturable microbes. Long-read sequencing technologies (e.g., Nanopore, PacBio) and single-cell genomics will improve genome assembly and characterization of challenging species. AI and machine learning will further optimize culture conditions and predict microbial interactions. These advancements will enhance the isolation, identification, and functional analysis of uncultivable microbes, supporting personalized microbiome-based interventions and therapeutic strategies. A well-established gut microbes library will advance our understanding of gut microbial dynamics and contribute to improved diagnostics, disease prevention, and treatment.