INTRODUCTION
Respiratory syncytial virus (RSV) is a 15 kb negative-sense, single-stranded RNA virus from the Paramyxoviridae family. It imposes a significant health burden globally, causing severe respiratory illness in young children, elderly populations, and individuals with chronic lung conditions. (1, 2, 3). Palivizumab and nirsevimab are mAbs for preventing RSV in infants and young children, with nirsevimab offering broader protection. Vaccines targeting the RSV preF protein—Abrysvo (for older adults and pregnant individuals to protect infants), Arexvy, and Mresvia (both for older adults)—are now approved. However, high costs, short half-life of mAbs, and limited antiviral treatment options emphasize the need for effective RSV-specific antiviral drugs (4). RSV infection begins in the upper respiratory tract through airway epithelial cells (AECs), which serve as the primary barrier against pathogens. Once RSV enters the nasal passages, it spreads to the lower respiratory tract, leading to viral replication and subsequent inflammation. This results in epithelial damage, including cell shedding, mucus overproduction, and compromised ciliary function, which contribute to the progression of bronchiolitis. The replication of RSV in both upper and lower airway epithelial cells further exacerbates disease symptoms (5, 6, 7). Understanding the function of airway epithelial cells is essential for advancing insights into immune responses, particularly given their role as the “gatekeeper” of severe RSV progression. Additionally, studies using in vitro models, such as upper and lower airway epithelial cells systems, have elucidated variations in antiviral immunity. Systems biology approaches, including transcriptional profiling, allow researchers to delineate virus-specific molecular pathways and immune responses. This is valuable for examining age-specific immune mechanisms and for comparing systemic and immune responses in RSV pathogenesis. Ultimately, these insights underscore the airway epithelium’s pivotal role in mediating immune defenses and hold promise for developing novel therapeutic strategies targeting the respiratory epithelium. This review discusses and summarizes the current state of knowledge on recent transcriptional profiling studies related to RSV infection, providing insights to enhance our understanding of RSV pathogenesis. Additionally, we have organized publicly available transcriptional profiling data on RSV infection into tables, categorizing studies by cell type or patient-based research in Tables 1 and 2. These systems biology approaches not only improve our understanding of RSV infection mechanisms but also pave the way for developing advanced antiviral strategies.
Table 1.
Transcriptome Datasets of RSV Infected Human Respiratory Cell Lines and Blood
| Title | Cells | Virus | moi | hpi | GEO accession | References |
|---|---|---|---|---|---|---|
| RSV directly binds host microRNAs and de-regulates host gene targets | A549 | RSV strain A2 | 0.1 | 48 | GSE231788 | (8) |
| Human RSV Non-Structural Protein NS1 enters the nucleus to disrupt gene transcription | A549 | RSV A2 strain | 1 | 96 | GSE155151 | (9) |
| Multiple RSV strains infecting HEp-2 and A549 cells reveal cell line-dependent differences in resistance to RSV infection | A549 | Four different RSV strains (A, B, GA1, GB1) | 0.01 | 24/48/72/96 | GSE196385 | (10) |
| Transcriptional response to SARS-CoV-2 infection | A549 | RSV A2 strain | 2 | 24 | GSE147507 | (11) |
| Dexamethasone inhibits RSV-driven mucus production while increasing viral replication without altering antiviral interferon signaling | NCI-H292 | RSV A2 strain | 0.5 | 48 | GSE140226 | (12) |
| Bromodomain Containing Protein 4 (BRD4) Regulates Inducible Expression of Its Interactome In The Innate Response To RSV | human small airway epithelial cells (hSAECs) | RSV Long | 1 | 24 | GSE179353 | (13) |
| Role of SWI/SNF-Related, Matrix Associated, Actin Dependent Regulator of Chromatin A4 (SMARCA4) core complex in epithelial innate response | human small airway epithelial cells (hSAECs) | RSV Long | 1 | 16/24 | GSE161846 | (14) |
| RSV-induced Cytoskeletal Inflammation | NHBE-ALI | RSV expressing green fluorescing protein (RSV-GFP), A2 | 4 | 144 | GSE146795 | (15) |
| Gene expression analysis of bronchial epithelial cells stimulated by different airway pathogens. | BEAS-2B | RSV A2 strain | 1 | 4 | GSE6802 | (16) |
| RSV gene expression | BEAS-2B | RSV Long | 1 | 4/24 | GSE3397 | (17) |
| IFNγ influences epithelial anti-viral responses via histone methylation of the RIG-I promote | AALEB cells (human bronchial epithelial cell line) | RSV A (M37) | 1 | 48 | GSE77154 | (18) |
| Network Analysis Reveals Age- and Virus-Specific Circuits in Nasal Epithelial Cells of Extremely Premature Infants | Nasal epithelial cells at the air-liquid interface (ALI) | RSV A | 1 | 48 | GSE239726 | (19) |
| Immune response to RSV in preterm and term infants | CBMC | RSV strain A2 and B | 1 | 24 | GSE196134 | (20) |
| DC response to RSV from adult peripheral and cord blood | blood samples (cord blood DC) | RSV A2 strain | 3 | 24 | GSE24132 | (21) |
| Analysis of cord blood B cells | Cord blood (CD19+Bcells) | RSV A | 5 | 6 | GSE78847 | (22) |
| Deciphering the gene expression response of human immune cells to stimulation with RSV in the context of antibodies | PBMC | RSV A2 | 1 | 24 | GSE59391 | (23) |
Table 2.
Publicly Accessible Transcriptome Datasets in RSV-Infected Patient-Based Research
| Title | N subjects |
Age demo -graphics | Sample type | Platform | GEO accession | References |
|---|---|---|---|---|---|---|
| Single-cell immune profiling reveals markers of emergency myelopoiesis that distinguish severe from mild RSV disease in infants |
212 RSV /56 controls | Infant | Blood (PAX gene) | Clariom GOScreen microarray | GSE246622 | (24) |
| Genome-wide analysis of whole blood transcriptional response to RSV, Influenza and Rhinovirus lower respiratory tract infection (LRTI) in children | 156 RSV /39 controls | Infant | Blood (Tempus) | Illumina Human HT-12 v4 | GSE38900 | (25) |
| Host transcriptome profile of children with respiratory virus infection |
29 mild /8 severe with RSV infection | children | Blood (PAX gene) | Illumina Hiseq 2500 | GSE155925 | (26) |
| The response of PBMCs and primary airway epithelial cells to Influenza and RSV virus |
51 RSV /10 controls | children | Blood (acid-citrate-dextrose) | Illumina HT-12 v3.0 |
GSE32140/ GSE32139/ GSE32138 | (27) |
| RSV disease severity: Insights into Protective Immune Responses |
89 RSV /34 controls | children | Blood (Tempus) | Illumina Human HT-12 v4 | GSE105450 | (28) |
| Host Gene Expression in Nasal and Blood Samples for the Diagnosis of Viral Respiratory Infection |
7 RSV /29 controls | children | Nasal, Blood (Tempus) | Affymetrix Human Clariom-D chips, | GSE117827 | (29) |
| Molecular Distance to Health Transcriptional Score and Disease Severity in Children Hospitalized with Community-Acquired Pneumonia |
19 RSV /17 controls | children | Blood (Tempus) | Illumina Human HT-12 v4 | GSE103119 | (30) |
| RSV Genotypes and Disease Severity in Young Children Hospitalized with Bronchiolitis |
49 RSV A /13 RSV B /17 controls | children | Blood (Tempus) | Illumina Human HT-12 v4 | GSE103842 | (31) |
| Host transcription profile in nasal epithelium and blood of hospitalized children under two years old with RSV infection | 106 RSV | children | Blood | Illumina Human HT-12 v4 | GSE97742/ GSE97741 | (32) |
| A simple screening approach to prioritize genes for functional analysis identifies a role for IRF7 in the control of RSV disease. | 27 RSV and 80 controls | children | Blood (PAX gene) | Illumina Human HT-12 v4 | GSE80179 | (33) |
| Nasopharyngeal microbiota, host transcriptome and disease severity in children with RSV infection | 106 RSV and 26 controls | children | Blood | Illumina Human HT-12 v4 | GSE77087 | (34) |
| Olfactomedin 4 serves as a marker for disease severity in pediatric RSV infection | 26 RSV | children | Blood | Affymetrix U133 plus 2.0 human microarrays | GSE69606 | (35) |
| Transcriptomic profiling in childhood H1N1/09 influenza reveals reduced expression of protein synthesis genes |
23 RSV /33 controls | children | Blood (PAX gene) | Illumina HT-12 v3.0 | GSE42026 | (36) |
| RNAseq of nasal cells from volunteers experimentally infected with RSV |
25 No cold /23 cold with RSV infection | Adult | nasal curettage tissue | Illumina Hiseq 2500 | GSE166161/ GSE155237 | (37) |
| Gene expression signatures of symptomatic respiratory viral infection in adults |
9 RSV /10 controls | Adult | Blood (PAX gene) | Affymetrix U133A human microarrays | GSE17156 | (38) |
GENE EXPRESSION PROFILING IN RSV INFECTED HUMAN CELL LINES
Human cell lines from the upper and lower respiratory tracts are commonly utilized in the investigation of RSV. Subsequent transcriptomic studies focus on the cellular responses elicited by RSV infection in these upper and lower airway cell lines (Fig. 1).
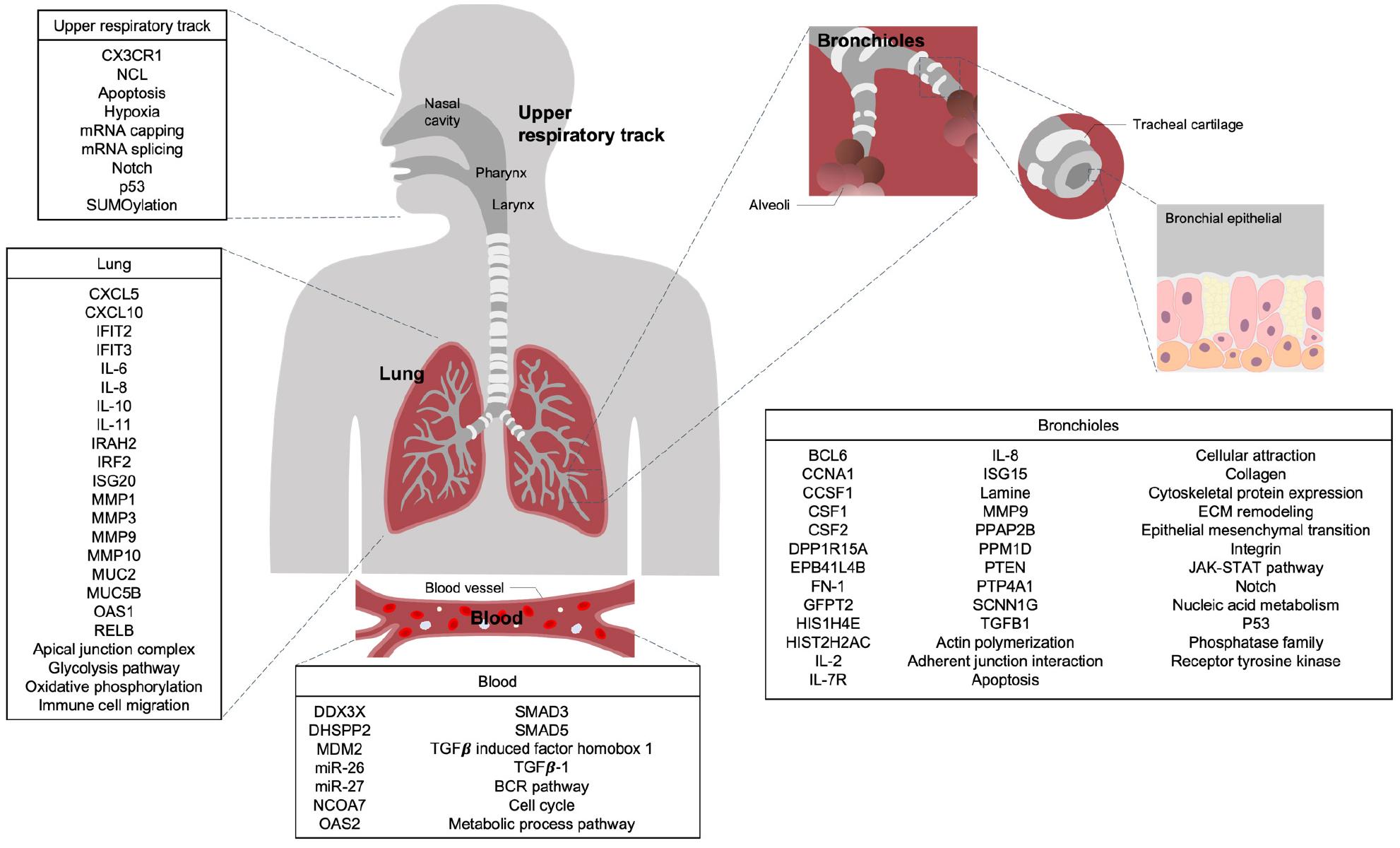
Fig. 1
Key Genes and Signaling Pathways Significantly Activated by RSV Infection in Human Respiratory Cell Lines and Blood. Based on transcriptional profiling studies, changes in key genes and major signaling pathways that are significantly upregulated following RSV infection, compared to uninfected controls, are shown for various cell types: nasal epithelial cells from the upper respiratory tract, A549 and NCI-H292 cells from the lung, hSAECs, NHBE-ALI, BEAS-2B, and AALEB cells from the bronchioles, and CBMC, cord blood-derived dendritic cells (DC), and cord blood CD19+ B cells from the blood.
Human lung cell lines
MicroRNAs (miRNAs) are critical regulators of nearly all physiological processes, yet their functions, particularly during viral infections, remain only partially understood. RSV binds directly to miR-26 and miR-27 through seed pairing. These miRNAs target different gene networks, with miR-27 focusing on cell cycle and metabolism, while miR-26 regulates antiviral immunity. Over the past two decades, RNA-RNA interactions between miRNAs and viruses have uncovered novel regulatory mechanisms by which miRNAs can function. Some viral-miRNA interactions have been shown to enhance genome stability and viral translation (39, 40, 41, 42). Although miRNA-target interactions have been identified in several viral families, they have been less explored in the A549 lung epithelial cell line, which is frequently used to model gene regulation during respiratory virus infections (42). To address this, a bioinformatics pipeline was developed to map miRNA-target interactions in RSV-infected and uninfected A549 cells. The analysis highlights miR-26’s involvement in the antiviral immune response to RSV, while miR-27 deregulation during infection affects key host genes linked to viral replication. Notably, miR-26 and miR-27 bind to specific sites in the RSV genome but are not degraded during infection. Gene Ontology (GO) analysis revealed no significant overlap between the pathways targeted by these miRNAs. MiR-27 targets genes associated with metabolism and cell cycle, such as Nuclear Receptor Coactivator 7 (NCOA7) and PH Domain And Leucine-Rich Repeat Protein Phosphatase 2 (PHLPP2), while miR-26 primarily targets genes involved in viral processes, including 2’-5’-Oligoadenylate Synthetase 2 (OAS2) and DEAD-box Helicase 3 X-linked (DDX3X). Upon RSV infection, miR-27 targets were not globally upregulated, although certain genes like Colony Stimulating Factor 1 (CSF1), involved in macrophage activation, showed increased expression (43). Conversely, miR-26 targets, such as Mouse Double Minute 2 Homolog (MDM2) and DDX3X, were consistently upregulated, aligning with its role in viral processes. The most significantly upregulated miR-27 targets, Cyclin G1 (CCNG1), PHLPP2, Golgi Membrane Protein 1 (GOLM1), and LIM Domain Kinase 1 (LIMK1), are critical for cell cycle arrest, IL-6 (Interleukin-6) regulation, and cytoskeletal remodeling, respectively (44, 45, 46, 47, 48). LIMK1 is particularly noteworthy, as inhibitors targeting it are currently under investigation as broad-spectrum antiviral therapies (49). miR-26 and miR-27 are key regulators of gene expression during RSV infection, modulating pathways critical for metabolism, cell cycle regulation, and antiviral immunity (8).
The C-terminal helix of RSV nonstructural protein 1 (NS1) plays a critical role in modulating gene transcription. Mutation of this region, particularly the Y125A substitution, results in an attenuated RSV strain with reduced expression of host immune response genes (50, 51, 52). RNA-seq analysis from A549 cells infected with either WT NS1 or the NS1 Y125A mutant at 96 hours post-infection (hpi) revealed significant differential expression of genes such as Interferon-Induced Protein with Tetratricopeptide Repeats 2 (IFIT2), Interferon-Induced Protein with Tetratricopeptide Repeats 3 (IFIT3), C-X-C Motif Chemokine Ligand 5 (CXCL5), Interleukin-1 Receptor-Associated Kinase 2 (IRAK2), Interferon Regulatory Factor 2 (IRF2), Interferon-Stimulated Gene 20 (ISG20), 2’-5’-Oligoadenylate Synthetase 1 (OAS1), and NF-KB Subunit (RELB) in WT infection, whereas the Y125A mutant showed minimal changes, resembling mock infection (9). A robust transcriptional response in A549 cells was observed at 72 and 96 hpi. While gene expression changes were minimal at 24 hpi, they rapidly increased after 48 hpi, reaching a peak of 6,000 to 8,000 upregulated and downregulated genes at 72 hpi. Gene set enrichment analysis (GSEA) revealed pathways associated with immune responses, cell cycle regulation, and metabolic processes, with divergent temporal dynamics. In A549 cells, the apoptotic response showed a consistent increase, peaking at 96 hpi. Cell cycle genes, including early region 2 binding factor (E2F) targets and G2M checkpoint genes, were upregulated at 48 and 72 hpi in A549 cells. Interferon-stimulated genes (IFN-α and IFN-γ) were strongly upregulated in A549 cells after 24 hpi. Cytokine signaling genes showed a steady increase in A549 cells, peaking at 96 hpi. Additionally, chemokine receptor genes displayed increasing activation through 96 hpi in A549 cells (10). RSV induces high expression of interferons (IFNs) and interferon-stimulated genes (ISGs), particularly at high multiplicity of infection (MOI) in A549 cells (11). An unbiased analysis of RNA sequencing data was performed to identify gene networks influenced by RSV and dexamethasone in RSV-infected NCI-H292 cells. Gene expression was assessed through gene ontology (GO) networks, revealing terms significantly upregulated by RSV and repressed by dexamethasone. The prominent GO terms associated with RSV infection and their repression by dexamethasone. Notably, while dexamethasone did not diminish antiviral and type I interferon-related GO terms, it significantly reduced pathways linked to immune cell migration and chemotaxis. The presence of several matrix metalloproteases (MMPs) suggests their involvement in the effects of dexamethasone on RSV infection and Muc5AC production (12).
Human bronchial cell lines
For the Human small airway epithelial cells (hSAECs), Bromodomain-containing protein 4 (BRD4) inhibition in airway epithelial cells alters alternative splicing, particularly in response to RSV infection. In the human small airway epithelial cells (hSAECs), treatment with the BRD4 inhibitor ZL0454 before and during RSV infection led to significant isoform switching in over 2000 genes and 3635 transcripts. RSV alone induced splicing events resulting in shorter open reading frames (ORFs), while BRD4 inhibition promoted longer ORFs and increased intron retention. Gene ontology analysis highlighted splicing changes linked to kinetochore assembly and nucleic acid metabolism, with 314 genes affected by both RSV and BRD4 inhibition (13). RSV-induced chromatin opening correlates with distinct gene expression profiles in hSAECs during infection. Using RNA-seq, increased chromatin accessibility was linked to gene upregulation, while reduced chromatin accessibility corresponded to gene downregulation. Pathway enrichment analysis revealed that receptor tyrosine kinase signaling, and extracellular matrix (ECM) organization were highly enriched, with key genes such as Glutamine-Fructose- 6-Phosphate Transaminase 2 (GFPT2), Transforming Growth Factor Beta 1 (TGFB1), JunB Proto-Oncogene, AP-1 Transcription Factor Subunit (JUNB), Fibronectin 1 (FN1), and Matrix Metalloproteinase 9 (MMP9) identified. These genes contribute to ECM remodeling and epithelial-mesenchymal transition (53, 54). Network analysis highlighted integrins, TGF, and cytoskeletal proteins as central hubs, underscoring the role of TGF-induced ECM-modifying proteins in RSV response (14). For Human bronchial epithelial cell line, RSV infection causes inflammation in the bronchiolar airways, leading to bronchial wall thickening, which is more severe in infants. The molecular mechanism of this thickening, particularly in adults, remains unclear. Using pseudostratified airway epithelium derived from human bronchial epithelial cells, researchers found that RSV primarily infects ciliated cells. These infected cells expanded significantly, contributing to increased airway epithelial height without compromising membrane integrity or ciliary function. This expansion is driven by cytoskeletal factors, such as Actin-Related Protein 2/3 (ARP2/3)-complex-driven actin polymerization, along with immunological factors like Interferon Gamma-Induced Protein 10 (IP10) / C-X-C Motif Chemokine Ligand 10 (CXCL10) and viral proteins such as Non-Structural Protein 2 (NS2). The phenomenon, termed cytoskeletal inflammation, reflects a noncanonical inflammatory response in RSV-infected cells, contributing to bronchial wall thickening. Infected epithelium demonstrated structural changes compared to mock-infected samples, including disorganized goblet cell granules, increased cell membrane folding (interdigitation), and a higher density of microvilli. Gene enrichment analysis, using the STRING database (a protein-protein interaction network database), revealed that most of the top enriched biological processes were related to cytoskeletal regulation, with a major cluster associated with actin filament organization. RSV significantly upregulated genes involved in actin polymerization, particularly through ARP2/3 complex-driven mechanisms (55, 56). Overall, RSV modulates actin cytoskeleton pathways, contributing to bronchial thickening and potentially revealing a novel mechanism of airway inflammation (15). RSV stimulation of Bronchial Epithelial Adenocarcinoma (BEAS-2B) cells revealed transcriptomic changes linked to transcription regulation, nucleic acid metabolism, and anti-apoptosis, reflecting the virus’s replication needs. RSV also induced inhibitory molecules, including dual specificity phosphatases such as Protein Phosphatase, Mg2+/Mn2+ Dependent 1D (PPM1D), Protein Phosphatase 1 Regulatory Subunit 15A (PPP1R15A), Protein Tyrosine Phosphatase Type IVA, Member 1 (PTP4A1), and Phosphatidic Acid Phosphatase Type 2B (PPAP2B), dampening innate immune responses. Despite dsRNA presence, RSV weakly triggered type I IFN-dependent genes through the TLR3-TRIF axis (16). Another study in RSV-infected BEAS-2B cells investigates the complex virus-host dynamics cells by analyzing gene expression. Using microarray, RSV-induced upregulation of arginase II (ARG2) protein was observed. Knockdown of ARG2 increased the release of Interleukin-8 (IL-8), Lactate Dehydrogenase (LDH), and histones, highlighting its role in RSV infection. Pathways such as P52 signaling, apoptosis, Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) signaling, and cytokine-cytokine receptor interactions were implicated. Early upregulation of cytokines Colony Stimulating Factor 1 (CSF1), Colony Stimulating Factor 2 (CSF2), and Interleukin-2 (IL2) was noted. Other pathways included ubiquitin-mediated proteolysis, cancer-related pathways, and cell cycle regulation. RSV also increased IL-8 and CSF2 release in a dose-dependent manner. Key genes— Epithelial Sodium Channel Subunit Gamma (SCNN1G), Erythrocyte Membrane Protein Band 4.1 Like 4B (EPB41L4B), CSF1, and Phosphatase and Tensin Homolog (PTEN)—were upregulated at both 4 and 24 hpi, while Tubulin Beta-1 Chain (TUBB1) was downregulated. Estrogen Receptor 2 (ESR2) showed a transient upregulation at 4 hpi, followed by downregulation at 24 hpi (17). Gene expression profiling via microarrays revealed innate immune genes potentially linked to the priming effect of interferon-gamma (IFN-γ) on respiratory epithelial cells following RSV infection. Notably, upregulation of antiviral genes Myxovirus Resistance 1 (MX1), Interferon-Induced Protein With Tetratricopeptide Repeats 1 (IFIT1), and IFIT3 was observed, highlighting the importance of Retinoic acid-inducible gene I (RIG-I) in viral detection and elimination (18).
Human upper respiratory tract cell lines
The immune response of nasal epithelial cells (NECs) to RSV in preterm infants is notably altered, characterized by diminished and imbalanced cytokine responses. Utilizing an air-liquid interface system that simulates in vivo conditions, this research reveals distinct differences in the innate receptor repertoire and antiviral responses between neonatal and adult NECs. Notably, C-X3-C motif chemokine receptor 1 (CX3CR1), linked to RSV, exhibits elevated expression in neonatal NECs, while neuroblastoma cell adhesion molecule (NCL) is more pronounced in adults. In adult NECs, NOTCH signaling is enriched following RSV engagement, indicating a differential response pathway compared to neonates. Additionally, RSV infection activates pathways related to mRNA capping, splicing, and SUMOylation, alongside triggering Toll-like receptor 2 and 2/6-dependent signaling, highlighting the complex age-related immune responses following RSV exposure (19).
Blood
To investigate the innate immune response to RSV in preterm and term infants, cord blood mononuclear cells (CBMCs) were stimulated with RSVA and RSVB (MOI = 1) for 24 hours. Analysis revealed 2846 overlapping genes between both groups, with functional enrichment showing cytokine–cytokine receptor interactions, RIG-1-like receptor signaling, and TLR signaling, indicating a conserved antiviral response. Term infants exhibited upregulation of 100 genes associated with cytokine signaling pathways, particularly IL-17, TNF, and IL-10 pathways. In contrast, preterm infants demonstrated reduced expression of critical antiviral genes, including Interleukin 10 (IL-10), X-C Motif Chemokine Ligand 1 (XCL1), CSF2, C-X-C Motif Chemokine Ligand 1 (CXCL1), C-X-C Motif Chemokine Ligand 2 (CXCL2), Interleukin 36 Gamma (IL-36γ), Suppressor of Cytokine Signaling 1 (SOCS1) and Suppressor of Cytokine Signaling 3 (SOCS3), with weaker immune coordination (20). RSV is a leading cause of morbidity and mortality, with T-cell responses potentially contributing to immunopathology via dendritic cells (DCs). Pediatric populations are particularly susceptible to severe RSV, yet DC responses in these groups remain underexplored. Primary DCs from cord blood and adult peripheral blood were compared after RSV infection. Transcriptomic analysis revealed differential regulation of the TGF-beta signaling pathway between age groups. TGF-beta1 (Transforming Growth Factor Beta 1) was reduced in adult DCs but elevated in cord blood DCs. Ingenuity Pathway Analysis identified TGF-beta family members, SMAD Family Member 3 (SMAD3), SMAD Family Member 5 (SMAD5), and TGF-b-induced factor homeobox 1 as key regulators in cord blood DC networks (21). RSV activates the B-cell receptor (BCR) pathway in natural regulatory B cells (nBreg cells). Cord blood nBreg cells, sorted as CD19+CD5+ CD10 B cells, were stimulated with RSV, leading to immunoglobulin M (IgM) secretion by IL-10-producing cells, indicating BCR involvement. Transcriptomic analysis showed that RSV and BCR stimulation induced similar gene expression profiles, with 534 genes upregulated, while Toll-like receptor 7/8 (TLR7/8) activation by the R848 agonist failed to mimic viral activation patterns. Pathway analysis confirmed significant upregulation of BCR-related pathways in RSV-activated nBreg cells, but not TLR, RIG-I, or CD40 pathways, highlighting BCR as a key mediator of RSV response in nBreg cells (22). RSV, both alone and in combination with human serum, triggers significant expression of innate inflammatory mediators. Microarray transcriptional profiling of adult PBMCs (peripheral blood mononuclear cells) revealed that inactivated RSV upregulated CXCL10 and C-X-C motif chemokine ligand 11 (CXCL11). The presence of human serum further amplified CXCL11 expression, while antibodies enhanced the transcriptional response of multiple interferon-alpha (IFNα) genes, underscoring the role of antibodies in modulating the innate immune response (23).
GENE EXPRESSION PROFILING IN RSV INFECTED PATIENTS
Numerous studies on RSV infection have been conducted through the analysis of transcriptomic profiles from infected patients across various age groups, aiming to observe age-specific cellular changes. Subsequent transcriptomic analyses explore the differential responses elicited by RSV infection in distinct age categories (Fig. 2).
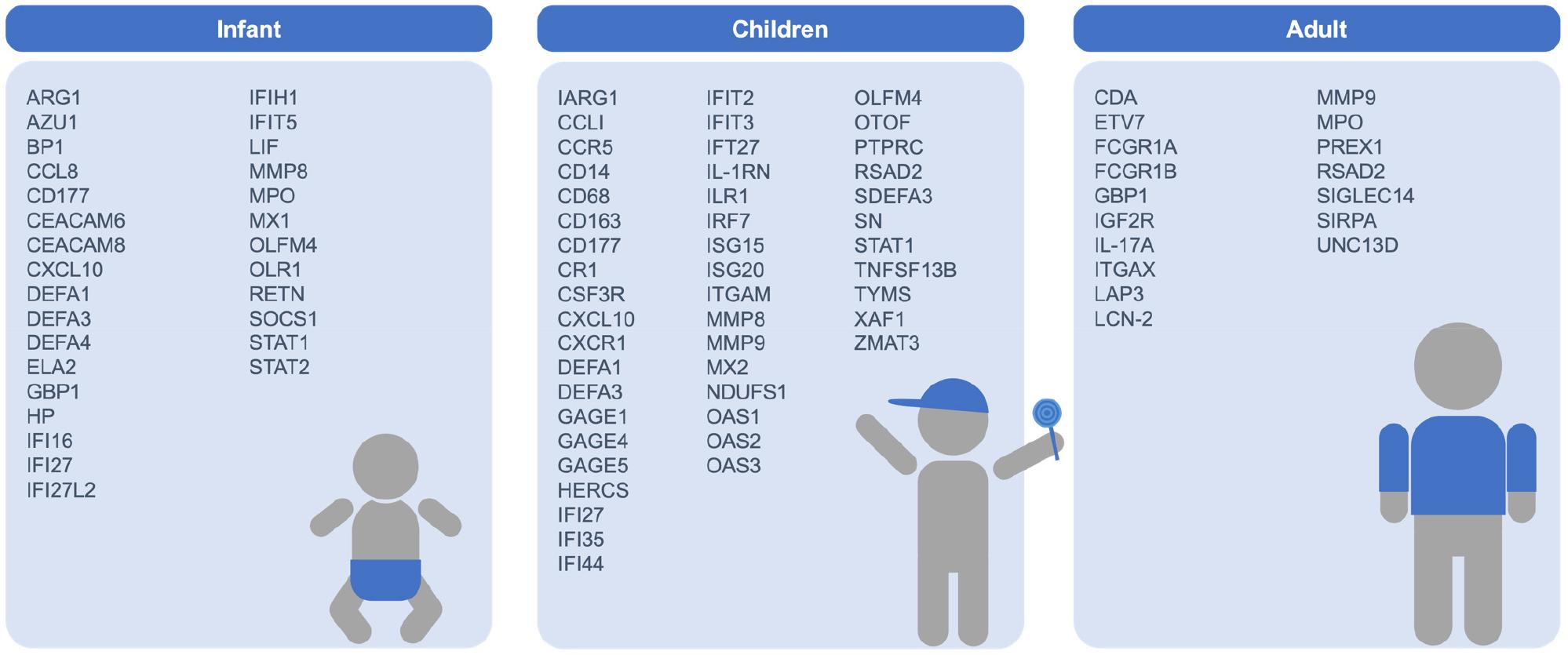
Fig. 2
Key Genes and Signaling Pathways Significantly Activated by RSV Infection in Human Respiratory Cell Lines and Blood. Based on transcriptional profiling studies, changes in key genes and major signaling pathways that are significantly upregulated following RSV infection, compared to uninfected controls, are shown for various cell types: nasal epithelial cells from the upper respiratory tract, A549 and NCI-H292 cells from the lung, hSAECs, NHBE-ALI, BEAS-2B, and AALEB cells from the bronchioles, and CBMC, cord blood-derived dendritic cells (DC), and cord blood CD19+ B cells from the blood.
Infant
In infants infected with RSV, differential expression analysis shows that mild RSV induces a strong IFN-driven antiviral response, while severe RSV upregulates genes related to myeloid leukocyte and neutrophil activation. GO pathway analysis reveals increased neutrophil-related gene expression in moderate and severe RSV, with lower expression in controls and severe cases. Overall, mild RSV activates an antiviral response, but severe disease shifts toward myeloid cell activation, as confirmed by whole blood transcriptomics (24). RSV infection induces a distinct transcriptional profile, with significant overexpression of neutrophil-related genes and suppression of B cell, T cell, and lymphoid lineage genes. Type I (IFIH1, IFIT1-5, STAT2, MX1) and type II interferon (IFI16, CXCL10, CCL8, GBP1-5, STAT1, SOCS1) genes were expressed solely in RSV cases. Neutrophil-related genes like Carcinoembryonic Antigen-Related Cell Adhesion Molecule 6 (CEACAM6), Defensin Alpha 4 (DEFA4), Myeloperoxidase (MPO), and Matrix Metallopeptidase 8 (MMP8) were highly overexpressed, while Defensin Alpha 1 (DEFA1), Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8 (CEACAM8), and Azurocidin 1 (AZU1) were exclusive to RSV. Notably, Lactotransferrin (LTF), Resistin (RETN), and Oxidized Low-Density Lipoprotein Receptor 1 (OLR1) were uniquely expressed in RSV infection, highlighting its specific immune signature (25).
Children
Differential gene expression analysis identified significant changes in blood samples of children with severe RSV compared to mild RSV. Upregulated genes included those associated with toll-like receptors, while downregulated genes were linked to transmembrane receptor tyrosine kinase signaling. Key genes, such as Cluster of Differentiation 177 (CD177), MMP9, and MMP8, showed increased expression in the RSV infection (26). Primary human airway epithelial cell (hAEC) cultures were inoculated with RSV, and total RNA was isolated for microarray analysis 48 hours post-infection. Differentially expressed genes (DEGs) identified included upregulated type I interferon-inducible genes such as interferon-induced protein 44 like (IFI44L), IFIT2, 2’,5’-oligoadenylate synthetase 3 (OAS3), myxovirus resistance 2 (MX2), interferon-induced protein 27 (IFI27), 2’,5’-oligoadenylate synthetase 1 (OAS1), interferon regulatory factor 7 (IRF7), signal transducer and activator of transcription 1 (STAT1), interferon-induced protein 35 (IFI35), and interferon-stimulated gene 20 (ISG20). Chemokines like CXCL10 and CCL5, cytokines such as TNFSF13B (tumor necrosis factor ligand superfamily member 13B), cell cycle-related gene XAF1 (X-linked inhibitor of apoptosis factor 1), and GAGE family members were also upregulated. Induced transcripts included CXCL10, oligoadenylate synthetase-like (OASL), IFI27, defensin alpha 1 (DEFA1), defensin alpha 3 (DEFA3), small nucleolar RNA (SN), and interleukin-1 receptor antagonist (IL-1RN), reflecting a robust host response during RSV infection (27). Children under 2 years were enrolled, including those with RSV infection and healthy controls. Differential expression analysis identified increased expression of shared transcripts associated with interferon, such as IFI27, OTOF (otoferlin), and IFIT3. In severe RSV cases, neutrophil-related genes like MMP9, DEFA1, and secretory defensin alpha 3 (sDEFA3) also exhibited increased expression (28). The analysis of biological processes and pathways was based on the upregulation of gene expression observed in nasal and blood samples, with heightened percentages in nasal samples due to RSV infections. Notably, cell cycle processes were uniquely enriched in RSV-infected subjects (29). The host immune response to different RSV types and genotypes was analyzed using blood transcriptional profiles from 62 infants with RSV bronchiolitis and 12 healthy controls. Of the infants with RSV LRTI, 79% had RSV A, and 21% had RSV B. Statistical analysis revealed 1,234 differentially expressed transcripts in RSV A cases compared to controls, and 640 for RSV B. Infants with RSV A exhibited higher interferon, inflammation, and neutrophil activity. Among genotypes, GA5 showed reduced interferon-related module activation but increased neutrophil response compared to GA2 and BA, indicating diverse immune modulation across RSV variants (31). RSV infection elicited potent, sustained innate immune responses, with consistent gene expression patterns across nasopharyngeal (NP) and blood samples. Interferon-α/β and NOTCH1 pathways, alongside biomarkers Histone Cluster 1 H4 Family Member E (HIST1H4E), Interleukin-7 Receptor (IL7R), Interferon-Stimulated Gene 15 (ISG15) in NP, and B-Cell Lymphoma 6 (BCL6), Histone Cluster 2 H2A Family Member C (HIST2H2AC), Cyclin A1 (CCNA1) in blood, were linked to RSV load and severity. BCL6 activation correlated with severe antiviral inflammation (32). The whole blood gene expression was analyzed in RSV-infected children and healthy controls, identifying significant differential expression. Key genes, including IRF7, OAS2, Radical S-Adenosyl Methionine Domain Containing 2 (RSAD2), HECT And RLD Domain Containing E3 Ubiquitin Protein Ligase 5 (HERC5), ISG15, IFI44, IL-1RN, Arginase 1 (ARG1), and IFIT3 were confirmed through network mapping analysis (33). Additionally, interactions between RSV and the nasopharyngeal microbiota modulate immune responses, potentially influencing disease severity and clinical outcomes (34). Microarray analyses identified Olfactomedin 4 (OLFM4) as a potential biomarker for disease severity in RSV-infected children. Comparison of PBMC from children with mild and severe disease revealed 564 differentially expressed probesets. A subsequent analysis of paired acute and recovery samples in severe cases identified 808 probesets. Notably, OLFM4 was the most upregulated gene (over 40-fold). Interestingly, apoptosis-related genes showed no significant upregulation, suggesting lymphopenia was not due to increased apoptosis (35).
Adult
In adults infected with RSV, susceptibility to RSV infection correlates with airway neutrophil activation. The baseline anti-RSV IgA titers were elevated in the No Cold group, though substantial overlap with the Cold group indicated that nasal IgA alone could not sufficiently account for susceptibility unless at extreme levels (57). Baseline RSV-specific CD8+ T cell frequencies in the lung did not differentiate between infected (Cold) and protected (No Cold) individuals, suggesting multifactorial correlates of protection (58). RNA sequencing of nasal tissue from both groups prior to inoculation identified 80 DEGs, with 73 (91%) higher in the Cold group, implicating neutrophil activation pathways as significantly enriched. The gene coexpression network analysis revealed a cluster of genes linked to neutrophil activity (59, 60). Correspondingly, nasal protein levels of neutrophil-associated mediators such as MPO, Lipocalin-2 (LCN-2) and IL-17A (Interleukin-17A), were significantly elevated in the Cold group at baseline, supporting the hypothesis that increased airway neutrophil activity enhances susceptibility to symptomatic RSV infection. No correlation was observed between baseline nasal antibody titers and neutrophil mediator levels, indicating independent susceptibility factors. Post-inoculation RNA-seq analysis at 3 dpi showed upregulation of DEGs, with significant immune-related pathways enriched in the No Cold group, including cytokine signaling and leukocyte migration, whereas the Cold group exhibited no significant enrichment in immune pathways. These results highlight the role of early inflammatory responses in protecting against symptomatic RSV infection (37). Peripheral blood was collected for whole-blood gene expression analysis at designated time points following intranasal viral inoculation. Gene expression was evaluated pre-inoculation and at peak symptoms for symptomatic individuals, while matched time points were used for asymptomatic individuals. This approach facilitated clustering based solely on expression patterns, reducing bias. RSAD2 (viperin) was identified as the most significantly differentially expressed gene in nasal epithelium 48 hours post-inoculation. A combined analysis of 84 time points yielded a single factor effectively distinguishing symptomatic from asymptomatic subjects, achieving 96.5% accuracy in leave-one-out cross-validation. Analysis of individual datasets confirmed that peak viral respiratory infection symptoms converge on a characteristic gene expression profile. Unique genes associated with RSV included Fc gamma receptor I (FCGR1A), Guanylate-binding protein 1 (GBP1), Leucine aminopeptidase 3 (LAP3), Ets variant 7 (ETV7), and FCGR1B, alongside key elements such as RSAD2, IFN response components, and the OAS gene family, reflecting the host response to viral infection (38).
CONCLUSION
We present a comprehensive summary of the current state of knowledge regarding publicly accessible data from transcriptional profiling studies on RSV infection, comparing gene expression changes in respiratory cell models and patient samples. This review focuses on enhanced gene expression analysis in upper and lower respiratory cell lines and age-specific responses in infants, children, and adults, with certain genes exhibiting significantly increased expression. We aimed to analyze the changes in gene expression induced by RSV infection, emphasizing the correlation between symptom development and RSV infection. Additionally, we examined gene expression patterns in patients to identify distinctions from those observed in infections with other respiratory RNA viruses, such as HRV and influenza virus. The findings presented in this review highlight that pathways associated with extracellular matrix (ECM) organization are notably activated in bronchiolar epithelial cells compared to other respiratory cells. In contrast, virus transport and B cell receptor signaling pathways exhibited distinct activation patterns in blood cell samples. In RSV-infected patient samples, infants demonstrated significant upregulation of genes such as RENT, OLFM4, and MMP8. Both OLFM4 and MMP8 are expressed in neutrophils, suggesting shared roles in neutrophil-mediated inflammation and tissue responses. Notably, OLFM4 serves as a potential marker gene for classifying disease severity in RSV infection and may regulate host innate immunity. It is also highly expressed in Th1 cells and is associated with immune response regulation during viral infections. In children, independent studies consistently reported increased expression of IRF7, DEFA1, CXCL10, IFI27, IFIT2, IFIT3, IL-1RN, and MMP9. These genes interact synergistically to regulate various aspects of the immune response during viral infection. IRF7 is critical for initiating viral infection signaling and modulates the expression of interferon-inducible genes, including CXCL10, IFI27, IFIT2, and IFIT3. IL-1RN and MMP9 are involved in regulating inflammation. These age-specific gene expression profiles suggest distinct roles in RSV pathogenesis for infants and children. Notably, genes such as MMP8, IFI27, STAT1, CXCL10, DEFA1, DEFA3, ARG1, and OLFM4 were upregulated in both groups. STAT1 induces the expression of IFI27 and CXCL10, directing immune cells to sites of inflammation. IFI27 inhibits viral replication and, together with CXCL10, amplifies immune responses. Furthermore, STAT1 is pivotal in the interferon signaling pathway, orchestrating the expression of IFI27 and CXCL10. ARG1 suppresses excessive inflammation and promotes tissue repair, while MMP8 and DEFA1/DEFA3 coordinate the inflammatory response. Collectively, these genes maintain a balanced immune response, regulating infection, inflammation, immune activity, and tissue repair. Comparisons between children and adults revealed similar upregulation of MMP9 and RSAD2 across both groups. RSAD2 exhibits antiviral properties through direct inhibition of viral replication and disruption of viral assembly processes. Meanwhile, GBP-1, which is activated by type I interferons during viral infections and plays a significant role in innate immunity, was consistently elevated in both infants and adults. Across all age groups, RSV infection prominently upregulated type I interferon and neutrophil activation pathways, highlighting their universal importance in the host immune response to RSV. These findings support the anatomical specificity of the respiratory tract environment to RSV infection, varying with age, thereby providing crucial insights into age-dependent viral pathogenesis. This review highlights respiratory cell-specific and age-specific increased transcriptional response patterns in RSV-infected patient samples, enhancing our understanding of RSV pathogenesis. These insights provide a foundation for developing targeted antiviral strategies to mitigate the target-specific and age-specific impacts of RSV infection.




