INTRODUCTION
Antimicrobial-resistant Staphylococcus aureus (S. aureus)is a worldwide concern due to its ability to develop resistance to multiple antimicrobials within a short period (1). S. aureus is a well-known opportunistic pathogen that colonizes the nasal cavity of humans and animals (2). Major virulence factors of pathogenic strains are hemolysin (α, β, γ, δ), leukocidin (Panton-Valentine leukocidin, PVL), enterotoxins (SEs), exfoliative toxins (ETA and ETB), and toxic shock syndrome toxin (TSST) (3). It infects both animals and humans causing infections ranging from superficial skin and soft tissue infections to life-threatening endocarditis, toxic shock syndrome and necrotizing pneumonia (4). Antimicrobials such as β-lactam, aminoglycoside, macrolides, tetracycline and quinolones are commonly used to treat staphylococcal infections (5, 6). Unfortunately, the level of resistance of these antimicrobials is increasing rapidly among staphylococci due to irrational use of antimicrobials in livestock production (5). Multidrug resistant (MDR) S. aureus is a major concern, which plays an important role in the transmission of resistance to different non- resistant bacteria through both mutation and/or the horizontal gene transfer (HGT) mechanism (7). Among β-lactam antimicrobials, penicillin, methicillin, cloxacillin, oxacillin, flucloxacillin, and dicloxacillin are the most useful antimicrobials to treat infections caused by S. aureus (7). Hence, methicillin-resistant S. aureus (MRSA) is a serious public health concern globally and is considered as one of the leading causes of hospital-acquired (nosocomial) infections in humans (8).In terms of mechanism, MRSA strains are resistant to all β-lactam antimicrobials by penicillin-binding protein (PBP2a) which is encoded by the mecA gene (6). Despite host specificity, human infections with animal originated S. aureus strains have been reported frequently (9). Livestock-associated MRSA (LA-MRSA) has been described to be capable of colonizing and infecting humans such as veterinarians and farmers because of their close contact with animals (10, 11). Vancomycin is a recommended drug for the treatment of MRSA infection; however, currently, the emergence of vancomycin resistance is a major public health concern around the world (12). VRSA strains are characterized by the expression of 11 van genes among which vanA gene is very common (12).
In Bangladesh, goat rearing is generally regarded as subsistence, smallholder and small-scale commercial operations. The majority of the farmers (80.5%) follow a semi-intensive system but few farmers (12.2%) use the free-range system while only 7.3% maintain a confinement pattern for goat production (13). Studies show wide variation in the prevalence of S. aureus in goats ranging from 16.7% to 96.2% in different countries (14). Several studies have reported the transmission of drug-resistant S. aureus (particularly MRSA and VRSA) from animals, environments, and humans worldwide (10, 11, 12). Similarly, antimicrobial-resistant S. aureus has also been reported in Bangladesh among poultry, cattle, and humans (7, 15, 16). However, despite being one of the most important food animals, data on drug-resistant S. aureus colonization in goats is sparse compared to other animal species in Bangladesh. Emphasis needs to be put on the investigation of antimicrobial-resistant S. aureus in small ruminants in order to develop effective prevention and treatment guidelines for S. aureus infections. This study was conducted aiming to understand the carriage frequency of S. aureus, spa type(s), antimicrobial resistance patterns, and virulence characteristics of S. aureus isolated from goats in Bangladesh.
MATERIALS AND METHODS
Ethical approval
This study was approved by the ethical committee of Chattogram Veterinary and Animal Sciences University, Bangladesh with retrospective effect from the date of its commencement. The memo no. is CVASU/Dir (R&E) EC/2021/273 (1).
Study population and sampling
This study was conducted in Chattogram, the second-largest city in Bangladesh. Sahedul Alam Quaderi Teaching Veterinary Hospital (SAQTVH) of Chattogram Veterinary and Animal Sciences University (CVASU) was selected as the sampling site because of its wide service coverage across the city. We selected random dates to visit the hospital and, on each visit, we randomly selected 1 to 10 goats for sampling. The nasal swabs were collected from goats by inserting a sterile swab into the nasal cavity followed by gentle rubbing of the nasal mucosa. Samples were then placed in a sterile 15 ml Falcon tube containing 5 ml Mueller Hinton broth (MHB) (Oxoid Ltd., UK) supplemented with 6.5% NaCl and transferred to the Microbiology and Veterinary Public Health Laboratory of CVASU for detailed investigation. The demographic and clinical data of animals were collected from the patient signalment sheets that were filled up by duty veterinarians.
Isolation and molecular identification of S. aureus
Swabs collected in MHB containing 6.5% NaCl were incubated at 37⁰C overnight for selective enrichment (17). After subsequent culture on 5% bovine blood agar, colonies showing hemolytic golden yellowish color were sub-cultured on Mannitol salt agar (Oxoid Ltd, UK). The phenotypic identification of presumptive staphylococci was done based on biochemical tests (18). Then, the primarily identified S. aureus was further confirmed by PCR targeting thermonuclease gene called nuc (19) which is species-specific and an indication of the pathogenic S. aureus. All PCR reactions were carried out in 25μl of a final volume containing 1μl DNA template (average concentration of 2.18ng/μl), forward and reverse primers of 1μl each (20 picomoles/μl), 12.5μl master mix (Thermo Fisher Scientific, Waltham, MA, USA) and 9.5μl nuclease- free water. MRSA ATCC 33591 strain and nuclease-free water were used as positive and negative controls, respectively.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of the S. aureus isolates was performed by the disk diffusion method (20) with a panel of eight antimicrobials from six different groups. Frequently prescribed antimicrobials in clinical settings along with the literature-reviewed less common alternatives to treat S. aureus infections were considered to design the panel. The antimicrobials with their respective class were: amoxicillin-clavulanic acid (30μg), cefoxitin (30μg), cefepime (30μg), tigecycline (15μg), vancomycin (30μg), sulfamethoxazole-trimethoprim (25μg), ciprofloxacin (5μg) and gentamicin (10μg) (Oxoid, UK). For each isolate, the zone of inhibition around each disk was measured and interpreted as susceptible (S), intermediate (I) or resistant (R) according to Clinical and Laboratory Standards Institute (CLSI) guidelines (21). S. aureus isolates showing resistance against at least three groups of antimicrobial agents (≥3) were defined as MDR (22).
Screening of MRSA and VRSA
Phenotypically, MRSA and VRSA were detected based on the CLSI guideline for cefoxitin (30μg) and vancomycin (30μg) disk diffusion with interpretive criteria of ≤ 21 and ≤ 16 mm inhibitory zone for methicillin resistance and vancomycin resistance, respectively (21). This phenotypic resistance was further investigated for the presence of the mecA and vanA gene by PCR as described earlier (23, 24). The primer sequences and PCR conditions are mentioned in Table 1. For mecA gene, positive and negative controls were used as mentioned earlier. For vanA gene, in-house isolated strain of vancomycin-resistant Enterococcus faecium and vancomycin-susceptible Enterococcus faecalis were used as positive and negative control, respectively.
Table 1.
Primer sequences of different genes used for the PCR in this study
| Gene | Primer name | Primer sequences (5′- 3′) |
Annealing Temp. | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| nuc | nuc-F | GCGATTGATGGTGATACGGTT | 55°C | 270 | Brakstad et al., 1992(19) |
| nuc-R | ACGCAAGCCTTGACGAACTAAAGC | ||||
| spa | spa-1113F | TAAAGACGATCCTTCGGTGAGC | 59°C | Variable | Kahl et al., 2005(25) |
| spa-1514R | CAGCAGTAGTGCCGTTTGCTT | ||||
| mecA | mecA P4 | TCCAGATTACAACTTCACCAGG | 55°C | 162 | Larsen et al., 2008(23) |
| mecA P7 | CCACTTCATATCTTGTAACG | ||||
| PVL | Luk-PV-1 | ATCATTAGGTAAAATGTCTGGACATGAT CCA | 55°/50°Ca | 433 | McClure et al., 2006(47) |
| Luk-PV-2 | GCATCAAGTGTATTGGATAGCAAAAGC | ||||
| sea | GSEAR-1 | GGTTATCAATGTGCGGGTGG | 57°C | 102 | Mehrotra et al., 2000(48) |
| GSEAR-2 | CGGCACTTTTTTCTCTTCGG | ||||
| seb | GSEBR-1 | GTATGGTGGTGTAACTGAGC | 57°C | 164 | Mehrotra et al., 2000(48) |
| GSEBR-2 | CCAAATAGTGACGAGTTAGG | ||||
| tst | GTSSTR-1 | ACCCCTGTTCCCTTATCATC | 57°C | 326 | Mehrotra et al., 2000(48) |
| GTSSTR-2 | TTTTCAGTATTTGTAACGCC | ||||
| vanA | vanA F | GGCAAGTCAGGTGAAGATG | 55°C | 713 | Azimian et al., 2012(24) |
| vanA R | ATCAAGCGGTCAATCAGTTC |
Detection of virulence genes
All confirmed S. aureus isolates were screened by PCR assay for the identification of predominant virulence factors; Panton-Valentine Leukocidin (pvl), enterotoxin (sea and seb), and toxic shock syndrome toxin (tst). The primer sequences and PCR conditions of these virulence genes are stated in Table 1.
S. aureus spa typing and phylogenetic analysis
For S. aureus protein A (spa) typing, firstly amplification of the polymorphic X region of the spa gene of the isolates was performed by PCR. The primers and cycling conditions used to amplify the spa gene were according to the methodology described earlier (25) (Table 1 ). The PCR product of the spa gene was purified using a DNA purification kit (Favorgen Biotech Corp, Taiwan) and then sequenced with the support of a commercial service (Macrogen Inc., Seoul, South Korea). The spa type of the isolates was assigned using Ridom Staph Type 1.4.1 software (Ridom GmbH, Würzburg, Germany) (26). Numerical spa repeat and type were assigned. The spa gene sequences of this study along with 13 other spa sequences obtained from BLASTN search results from NCBI GenBank were aligned using clustalW in MegaX. The Maximum Likelihood method with a bootstrap value of 1000 replicates was applied to construct the phylogenetic tree.
Statistical analysis
All data were entered and analyzed using the “R” program (version 3.5.1). The univariate logistic regression analysis was performed to assess the relative association between the presence of S. aureus and different demographic factors and clinical manifestations. The heat-map depicting the distribution of antimicrobial resistance phenotype and genotype was prepared by using GraphPad Prism 7 (La Jolla, CA, USA). Variable(s) having Pof ≤0.05 was considered as statistically significant.
RESULTS
S. aureus isolates
Out of the 200 animals investigated, 23 (11.5%; 95% CI 8-17%) tested positive for S. aureus based on the presence of the nuc gene (Fig. 1a).The univariate logistic regression analysis showed that the percentage of S. aureus was higher in goats with cutaneous and mucosal lesions than in goats without such lesions (P=0.01) and it varied significantly across the goat breeds (P= 0.02) (Table 2).

Fig. 1
Result of PCR assay for detection of Staphylococcus aureus (nuc, spa), antimicrobial resistance genes (mecA, vanA) and virulence gene (sea).
Table 2.
Risk factors associated with prevalence of S. aureus in goats
Antibiogram profiles of S. aureus isolates
The antimicrobial susceptibility pattern of 23 S. aureus isolates showed that all the isolates tested were resistant to ciprofloxacin (100%) followed by cefoxitin (74%), amoxicillin-clavulanic acid (65%), sulfamethoxazole-trimethoprim (65%), vancomycin (57%), cefepime (57%) and tigecycline (39%). On the contrary, the majority of the isolates showed sensitivity to gentamicin (78%) (Table 3). Individual antibiogram profiles of all isolates are illustrated in Fig. 2. Of all, 21 (91%) isolates were found as MDR. The MDR pattern of the isolates has been summarized in Table 3.
Table 3.
Antimicrobial susceptibility profiles of S. aureus isolates (n= 23)
Detection of resistance genes
Among phenotypically 17 cefoxitin-resistant isolates, only one was found positive for the mecA gene to be considered as MRSA (6%; 95% CI 0-29%) (Fig. 1b). Therefore, the remaining 22 isolates were methicillin-susceptible (MSSA). In addition, among 13 phenotypically vancomycin-resistant S. aureus isolates, 3 (23%; 95% CI 8-51%) were positive for vanA gene detected in this study (Fig. 1c).
Virulence attributes of S. aureus
Out of the virulence factors investigated in the 23 staphylococcal isolates, four (17%; 95% CI 6-38%) were positive for the sea gene (Fig. 1d) that encodes enterotoxin A, one of them was MRSA. No isolates tested positive in the PCR test for seb, tst and PVL gene.
spa types and phylogenetics
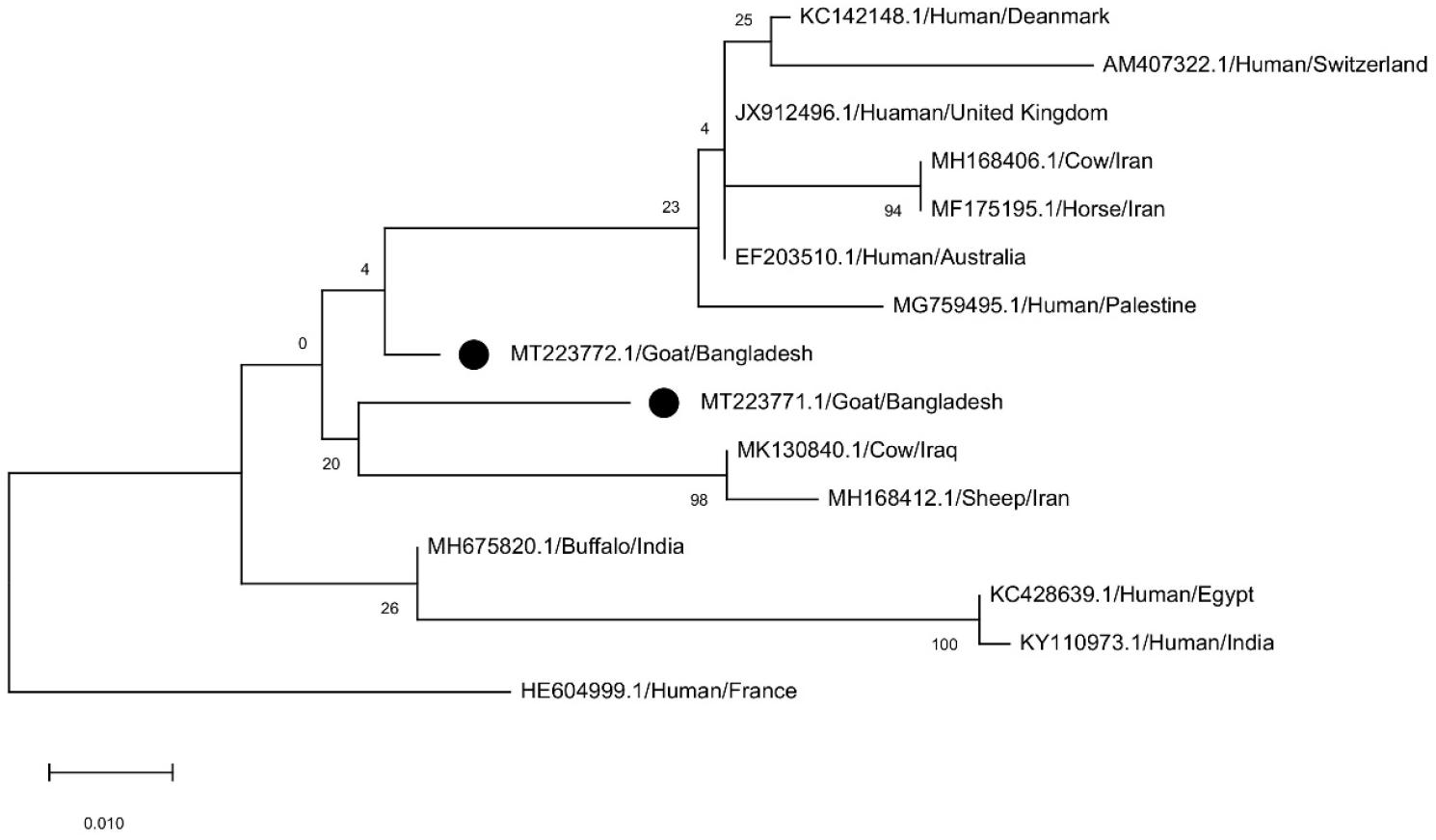
Amplification of polymorphic X region of the spa gene resulted in the formation of two different DNA bands with the sizes of ~ 300 to 400 bp (Fig. 1e), then we sequence one from each pattern as a representative. One of them was MRSA strain and another one was MSSA. The spa gene sequences were submitted to the NCBI GenBank (accession MT223771 and MT223772). The spa gene sequence (MT223771) of MSSA isolate was typed as t5259and another sequence (MT223772) of MRSA appeared to be novel, as it was not described yet in the spa Ridom Server used for analysis in this study. The repeat successions for these two sequences were 26-23-17-34-17-12-16 and 07-23-12-21-12-41-13-17-12-12-17 respectively. The Kreiswirth IDs of them were T1J1M1B1M1G1K1 and U1J1G1F1G1U2E1M1G1G1M1, respectively. The number of repeat units was 7 and 11, consecutively whereas the lengths of the entire VNTR were 168 and 264, respectively. For t5259, the repeat sequence started at 392 and for the untyped one, it started at 77. Phylogenetic analysis showed greater similarity between these two spa sequences (Fig. 3).
DISCUSSION
Studies have shown that goats with or without clinical condition(s) like respiratory illness can harbor S. aureus in their nares (4, 5). A wide variation in the prevalence estimate of S. aureus from goats has been reported which ranges from 5.6% to 43.2% or even more (27, 28, 29). Here, we reported the percentage of S. aureus as 11.5% in goats attending a teaching veterinary hospital, in Bangladesh.
S. aureus is widely resistant to the β-lactam group of antimicrobials which is evident in this study as well (1, 27). Almost 57% of isolates in this study were vancomycin-resistant (VRSA) as revealed by the disk diffusion method, which is the drug of choice for the treatment of MRSA infections though this antimicrobial is not commonly prescribed in goat practices in Bangladesh (30). Out of 13 phenotypically vancomycin-resistant isolates, 3(23%) were positive for the vanA gene which is lower than the previous study findings from other Asian countries (12). The remaining isolates might possess other van genes that were not tested in this study. However, heteroresistance might be responsible for seeing such a high rate of vancomycin resistance in S. aureus(31). Surprisingly, about 39% of isolates were resistant to the first glycylcycline antibiotic, tigecycline, which is regarded as an effective therapy against MSSA and MRSA without any co-resistance mechanisms yet to be known (32). Approximately, 91% of isolates were MDR. Selective pressure imposed by the use of antimicrobials as therapeutic or chemoprophylaxis might be the main cause of seeing such an unprecedented rate of multidrug resistance among S. aureus isolates (33). Isolation of MRSA circulating in the goat population has great public health significance since LA-MRSA can be transmitted to humans, especially to farmers, slaughter-house workers, and consumers of small-ruminant products in general (34) due to direct contact with animal, and can cause severe infection (9, 10, 11). In this study, 74% of isolates have shown resistance to cefoxitin, among which only one (6%) isolate was found to carry mecA gene confirming it as a methicillin-resistant isolate.
We acknowledge that due to lack of logistics, we studied only mecA gene for MRSA identification (which is considered as gold standard), we did not study mecC gene (a novel mecA homologue) as suggested for definitive identification of MRSA (35). However, marked variation in the prevalence of MRSA based on geography, host, environmental settings, etc. has already been described across Europe, Asia, and the USA (1, 27, 28, 32, 36). Among the virulence factors investigated, 4/23 strains were positive for the sea gene which is one of the classical enterotoxin producing genes. Although nine different genes are responsible for heat-stable staphylococcal enterotoxins (SEs), still the sea is considered the most prevalent one (37, 38) with occasional exceptions (3, 28). Epidemiological studies in humans suggested that the majority of S. aureus-associated infections and outbreaks have been caused by isolates with the sea type enterotoxin (39, 40). Notably, two strains were positive for both the sea gene and coagulase enzyme indicating high pathogenic potential.
A novel spa type from an MRSA isolate, not yet described in Ridom spa Server, was detected in this study. Another spa type, which is t5259, detected in this study was also rarely seen in the S. aureus isolates of small ruminants. A study in Japan reported the same spa type t5259 as MSSA from dairy cattle (41). The predominant spa types of S. aureus from goats in Europe are t1166 and t318 (42) and from sheep t002, t1534, t2678 and t3576 (43). There are few reports on the spa typing of S. aureus isolates in Bangladesh. One study involving S. aureus of ready-to-eat foods origin in Dhaka, Bangladesh identified seven different spa types with the predominance of t1198 and t315 (44). Notably, the novel spa type identified from this study was also an MRSA strain. Our study suggests that the carriage frequency of S. aureus was highest in goats with cutaneous and mucosal lesions such as dermatitis and oral lesions. Several previous studies indicated that animals with chronic infections such as dermatitis, and otitis require veterinary interventions and antimicrobial therapies more frequently which, in turn, increases the transmission burden of nosocomial pathogens like S. aureus and also their high antimicrobial resistance (45, 46).
In conclusion, this study has reported the high frequency of MDR S. aureus from goats in Bangladesh. Detection of vanA gene indicates the dissemination of vancomycin resistance among S. aureus limiting the treatment options. Since human transmission with a particular goat-originated MRSA strain is possible, a systematic study covering the whole country should be conducted coupled with an awareness campaign among livestock-associated personnel.