INTRODUCTION
Mast cells (MCs) serve as sentinel cells residing strategically at the interface between the host and the external environment in organs such as the skin, airways, gastrointestinal tract, and urinary tract. While historically recognized for their pivotal role in triggering asthma and allergic reactions through the release of histamine and other mediators (1), emerging evidence is reshaping our understanding of MCs.
Beyond their involvement in allergic responses, these multifunctional cells have been increasingly recognized for their crucial roles in immune surveillance and host defense. Upon encountering pathogens, MCs initiate a multifaceted response involving the release of an array of granule contents. These include not only histamine and heparin, but also proteases, antimicrobial peptides, and cytokines. This orchestrated response contributes to pathogen eradication through direct elimination and the recruitment of immune cells, thereby initiating effective inflammatory responses (2, 3). Given their prominent localization within barrier tissues, the regulated functioning of MCs is of critical importance to sustain tissue homeostasis and overall health. Consequently, deciphering the intricate mechanisms that underpin the multifunctional nature of MCs as orchestrators of immune equilibrium is essential.
Macrophages, representing a cornerstone of innate immune defenses, are ubiquitously distributed across tissues and hold multifaceted roles in immune responses, wound healing, and the mediation of inflammation (4, 5). The apparent dichotomy in macrophage functions stems from their remarkable transcriptional plasticity. This adaptability allows macrophages to adopt distinct profiles contingent upon the surrounding microenvironment (6). Macrophages activated by interferon (IFN)-γ or pathogen-associated molecular patterns, such as lipopolysaccharide (LPS), undergo a polarization process leading to the M1 phenotype. This polarization induces the synthesis of proinflammatory cytokines, as well as reactive oxygen and nitrogen species, thereby driving microbicidal activities. In contrast, macrophages exposed to Th2 cytokines or immune regulatory cytokines, such as transforming growth factor-β, shift toward the M2 phenotype. M2 macrophages actively suppress inflammation, promote tissue regeneration, and enhance homeostatic responses (7).
Recent investigations have unveiled the capability of MCs to generate inflammatory mediators that induce macrophage activation and proliferation within inflammatory microenvironments (8). However, the question of whether MCs exert regulatory control over macrophage polarization under normal physiological conditions remains elusive. In this study, we investigated the intricate interplay between MCs and macrophages, exploring their impact on macrophage polarization and transition. By employing an in vitro approach simulating conditions of normal homeostasis, we aimed to elucidate the potential mechanisms through which MCs modulate macrophage polarization. Additionally, we propose factors that could be involved in MC-mediated macrophage polarization, contributing to a deeper understanding of the interplay between these immune cell populations and their collaborative roles in maintaining immune homeostasis.
MATERIALS AND METHODS
Cell culture
The human MC line HMC-1 was kindly provided by Dr. Ji Young Kim (9) from the Radiation Health Institute, Korea Hydro & Nuclear Power Co., Ltd., Seoul, South Korea. HMC-1 cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 U/mL penicillin.
THP-1 cells (ATCC, Manassas, VA, USA), a human monocyte cell line, were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (Welgene, Daegu, Korea) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin solution, and 50 μM 2-mercaptoethanol.
Generation of conditioned medium from HMC-1 cell cultures (HMC-1 CM)
To generate HMC-1 CM, 107 HMC-1 cells were cultured in 10 mL serum-free IMDM for 48 h. Subsequently, the medium was collected and centrifuged at 190 × g for 5 min at 25°C. The supernatant was passed through a 0.2µm filter (Pall Corporation, Port Washington, NY, USA) and centrifuged at 5000 x g for 1h at 4°C. (Sorvall LYNX4000, Thermo Fisher Scientific, Waltham, MA, USA). The concentrated CM (10fold) was stored at −80°C for future use. As a negative control, serum-free culture medium underwent the same processing steps.
THP-1 polarization
After an initial 6-h stimulation with 150 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Saint Louis, MO, USA), the THP-1 cells (4 Χ 105 cells per well in 12 well plates) were subjected to an additional 48-h incubation to induce polarization toward either the M1 or M2 phenotype. For M1 polarization, the culture medium was supplemented with INF-γ (20 ng/mL, Peprotech, Cranbury, NJ, USA) and LPS (100 ng/mL, Sigma-Aldrich). Conversely, M2 polarization was induced with IL-4 (20 ng/mL, Peprotech) and IL-13 (20 ng/mL, Peprotech). Both polarization conditions were in the presence of PMA. Cells cultured for 48 h in the presence of PMA without additional stimuli were used as M0 macrophages. Morphological changes were observed utilizing a phase-contrast microscope. To assess the impact of HMC-1 CM on THP-1 polarization, 50 uL of HMC-1 CM was introduced to total 1.5 mL culture media along with the polarization-inducing cytokines. Additionally, the tryptase inhibitor, APC 366 (20μg/ml, Sigma–Aldrich), was used to confirm the effect of tryptase in M1 polarization.
Quantitative reverse-transcription PCR (qRT-PCR)
To validate the expression of macrophage makers, M0, M1, and M2 macrophages were harvested, followed by RNA extraction using TRIzol (Invitrogen, Waltham, MA, USA). Complementary DNA was synthesized using a reverse transcription reagent (ELPIS-Biotech, Daejeon, Korea) according to the manufacturer’s instructions. qRT-PCR analysis was conducted using a StepOnePlus instrument (Applied Biosystems, Foster City, CA, USA) and a SensiFAST SYBR Hi-ROX kit (Bioline, London, UK). All gene expression values (Inos, Tnfa, Il1b, Il6, Il23, Arg1, Tgfb, Il1ra, Cd206, and Il10) were normalized to the Gapdh reference gene. Primers utilized in this study are provided in Table 1.
Table 1.
qRT-PCR Primers
Flow cytometry
To validate the expression of cell-surface antigens, macrophages were subjected to staining with FITC-conjugated anti-human CD206 antibody (Biolegend, San Diego, CA, USA) and PE-conjugated anti-human CD284 (TLR4) antibody (Biolegend). Expression levels were measured using a NovoCyte flow cytometer (ACEA Biosciences, San Diego, CA, USA) and analyzed using NovoExpress software. To assess non-specific binding, staining was also performed using FITC-conjugated isotype-control mouse IgG1 (Biolegend) and PE-conjugated mouse IgG2a (Biolegend).
Enzyme-linked immunosorbent assay (ELISA)
For the quantification of tryptase levels in HMC-1 CM, a Human Tryptase ELISA kit was used according to the manufacturer’s instructions (Innovative Research, Novi, MI, USA). As a control, conditioned medium from THP-1 cells was similarly measured.
Statistical analysis
All presented data are shown as mean ± standard error of the mean (SEM). Statistical significance was determined using one-way ANOVA or two-way ANOVA, facilitated by GraphPad PRISM 10 software (GraphPad Software, San Diego, CA, USA). A P-value of less than 0.05 (P<0.05) was considered statistically significant.
RESULTS
THP-1 monocytes were successfully polarized into M1 and M2 macrophages
Human THP-1 monocytes were successfully differentiated into M0 macrophages and polarized into distinctive M1-like macrophage and M2-like macrophage phenotypes upon stimulation. This encompassed an initial 6-h incubation in the presence of 150 ng/ml PMA, followed by a 48-h incubation with additional specific skewing cytokines necessary for the polarization to M1-like or M2-like macrophages, following a previous protocol with slight modifications (10).
Microscopic examination of the resultant macrophage populations revealed distinct morphological differences corresponding to each polarization state. Unpolarized macrophages (M0), treated solely with PMA, adhered to the surface without displaying specific morphological characteristics. In contrast, M1-like macrophages exhibited pronounced morphological features characteristic of classically activated macrophages, including an irregular, elongated shape with protrusions, indicative of their heightened activation state. On the other hand, M2-like macrophages displayed a more rounded and less pronounced morphology (Fig. 1A).
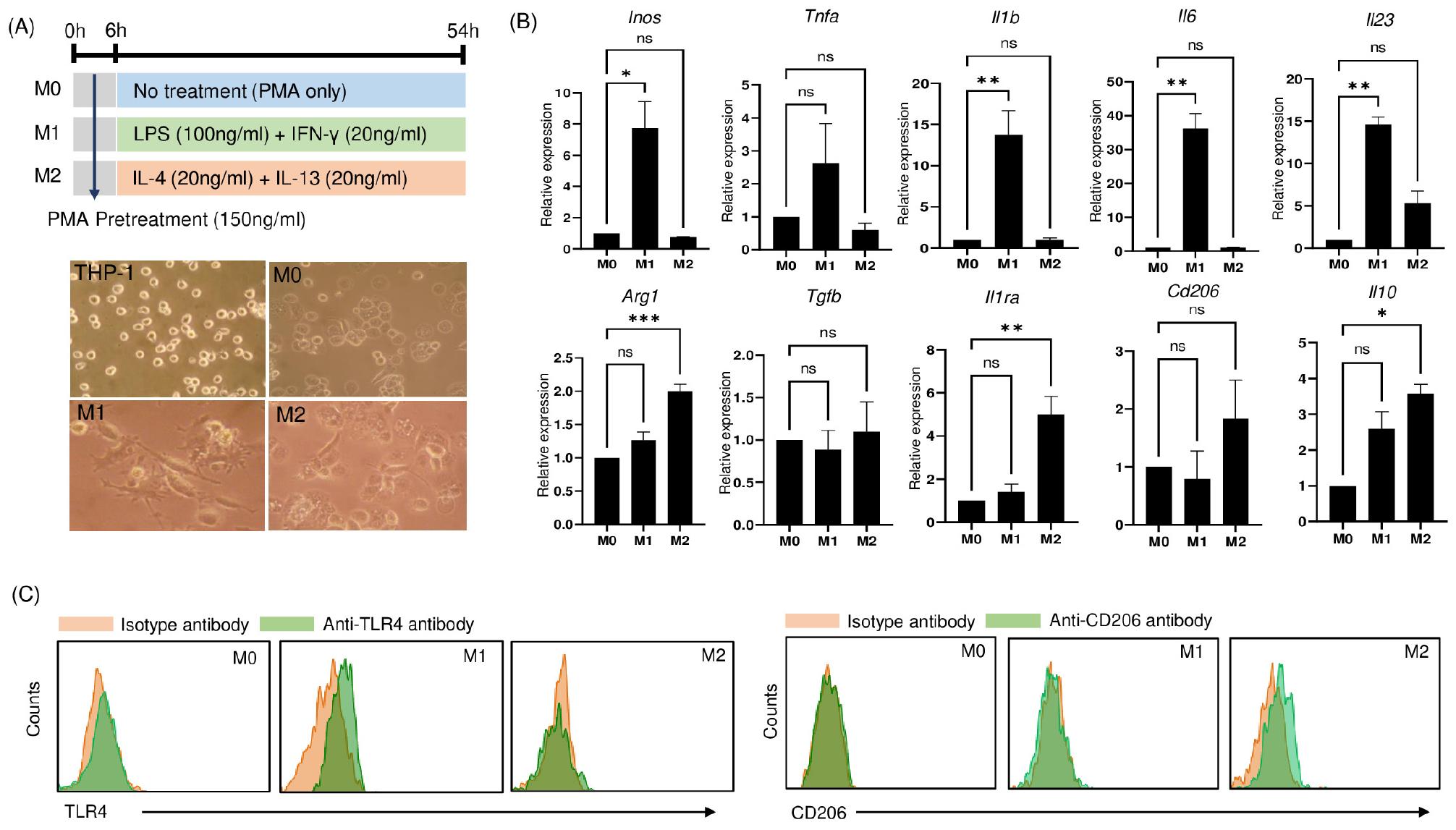
Fig. 1
Phenotypic characterization of differentiated THP-1 cells. (A) Experimental schematic illustrating the differentiation and polarizing processes of THP-1 cells into M0 macrophages, M1-like macrophages, and M2-like macrophages using specific stimuli (upper). Lower is representative microscopic images of THP-1 monocytes differentiated into non-polarized M0 macrophages (PMA alone), classically activated M1-like macrophages (further stimulated with IFN-γ/LPS), and alternatively activated M2-like macrophages (further stimulated with IL-4/IL-13). Phase-contrast images were captured at 200× magnification. (B) qRT-PCR analysis of various markers, including M1 markers (Inos, Tnfa, Il1b, Il6, and Il23) and M2 markers (Arg1, Tgfb, Il1ra, Cd206, and Il10) in polarized macrophages. Data are presented as mean ± SEM and analyzed by Ordinary one‑way ANOVA with Dunnett’s multiple comparison test (*P<0.05, **P<0.01, ***P<0.001). (C) Flow cytometric analysis depicting surface expression of TLR4 (M1 marker) and CD206 (M2 marker) in polarized macrophages.
Moreover, an in-depth assessment of gene expression patterns sheds light on the transcriptional dynamics of these polarized macrophages. M1-like macrophages displayed a significant upregulation of transcripts linked to proinflammatory responses, including Inos, Tnfa, Il1b, Il6, and Il23 (Fig. 1B). In contrast, M2-like macrophages exhibited a distinct gene expression profile characterized by elevated levels of Arg1, Tgfb, Il1ra, Cd206, and Il10, which are markers associated with the M2 phenotype.
Flow cytometry analysis provided further confirmation of the distinct features of the polarized macrophage subsets. M1-like macrophages displayed induction of TLR4, while CD206 induction was a distinguishing characteristic of M2-like macrophages, both in comparison to unpolarized M0 macrophages (Fig. 1C).
HMC-1 CM modulates macrophage polarization to M1-like phenotype
We further explored the potential of HMC-1 cells to influence macrophage polarization by supplementing HMC-1 CM with skewing cytokines during the initiation of macrophage polarization.
Upon comparing the expression profiles of M1 and M2 markers within the macrophage groups (M0, M1, and M2) following treatment with HMC-1 CM, intriguing patterns emerged. M0 macrophages treated with HMC-1 CM displayed a significant increase in M1 markers, including Inos, Tnfa, Il1b, and Il6, suggesting a propensity toward M1-like polarization. In contrast, there were no significant differences in the expression of M1 markers in either M1-like macrophages or M2-like macrophages treated with HMC-1 CM, indicating a different response to HMC-1 CM in these cells compared to M0 macrophages (Fig. 2A).
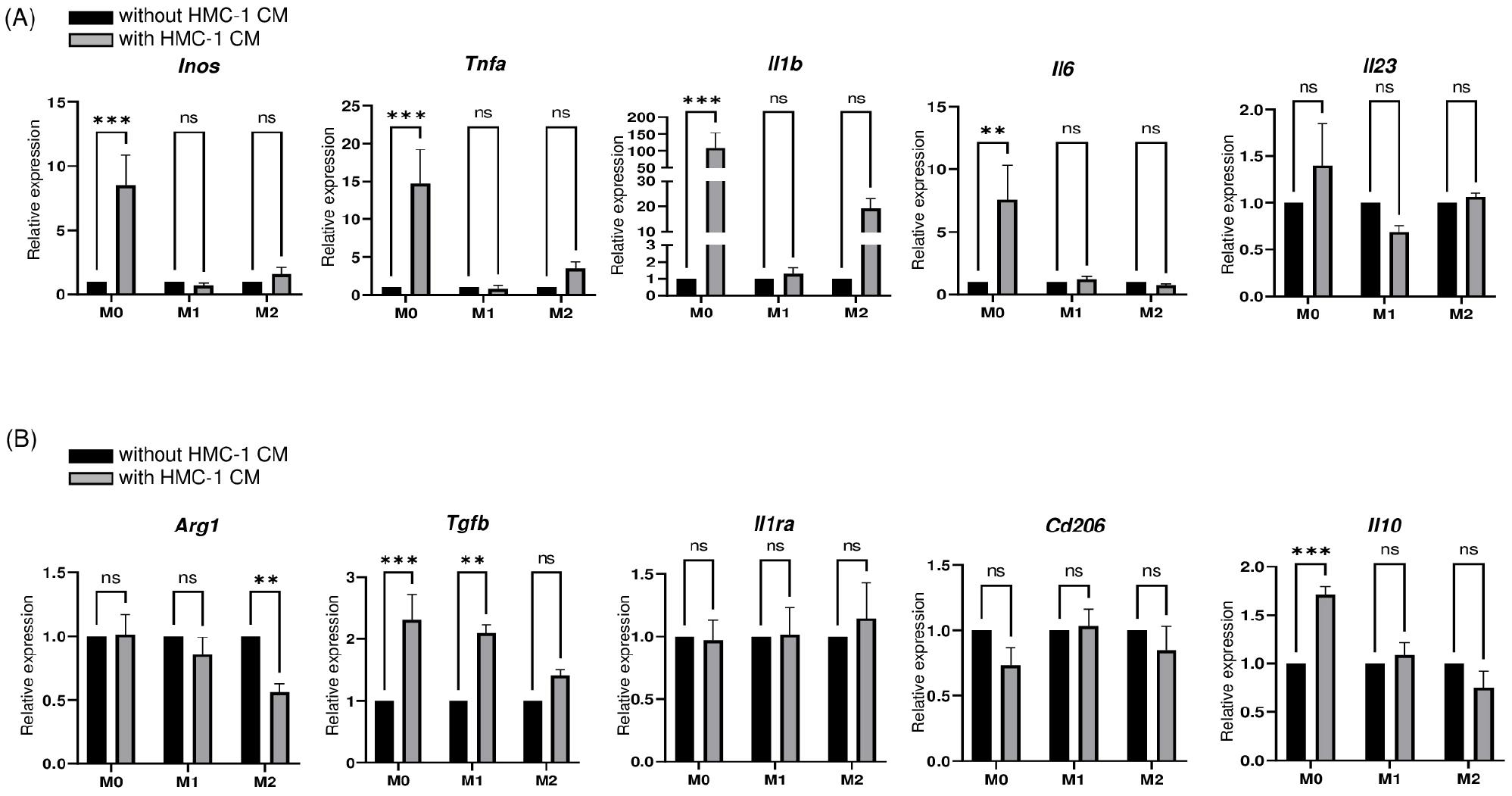
Fig. 2
Co-treatment with PMA and HMC-1 CM promotes M1-like macrophages polarization. (A) qRT-PCR analysis of M1 markers, including Inos, Tnfa, Il1b, Il6, and Il23, in macrophages co-treated with or without PMA and HMC-1 CM. Data are presented as mean ± SEM and analyzed by two-way ANOVA with Sidak’s multiple comparison test (**P<0.01, ***P<0.001). (B) qRT-PCR analysis of M2 markers, including Arg1, Tgfb, Il1ra, Cd206, and Il10, in macrophages co-treated with or without PMA and HMC-1 CM. Data are presented as mean ± SEM and analyzed by two-way ANOVA with Sidak’s multiple comparison test (**P<0.01, ***P<0.001).
When examining M2 markers, the presence of HMC-1 CM induced distinct effects. The expression of Arg1 was significantly decreased in M2-like macrophages, while Il1ra and Cd206 expression remained largely unaffected. However, Tgfb and Il10 showed significantly higher expression levels in M0 macrophages upon exposure to HMC-1 CM. Interestingly, even in M1 -like macrophages, the addition of HMC-1 CM led to a significant increase in Tgfb expression (Fig. 2B). Taken together, our findings indicate HMC-1 CM promotes a polarization preference toward the M1-like phenotype over the M2-like phenotype. However, this effect appears to be subject to modulation, as suggested by the induction of Tgfb and Il10 expression.
HMC-1 CM augments M1-like phenotype in M2-like macrophages
The observed propensity of HMC-1 CM to induce M1-like polarization prompted us to investigate its impact on the plasticity between macrophage phenotypes in cells that had already undergone polarization. Thus, we added HMC-1 CM to macrophages that were previously stimulated with skewing cytokines for 48 h. Subsequently, we continued to culture these macrophages for an additional day and assessed the resultant expression profiles of M1 and M2 markers.
The addition of HMC-1 CM yielded a modest increase in Inos, Il1b, Il6, and Il23 expression in M2-like macrophages (Fig. 3A). The extent of polarization toward M1-like phenotype observed in M0 macrophages was less pronounced compared to when HMC-1 CM was initially added during the polarization culture (Fig. 2). However, Il1b expression remained significantly elevated, indicating a sustained HMC-1 CM stimulus is necessary for robust M0-to-M1-like polarization. The expression of M2 markers demonstrated minimal changes in response to HMC-1 CM, except for Cd206, which exhibited a modest but significant increase in M2-like macrophages (Fig. 3B).
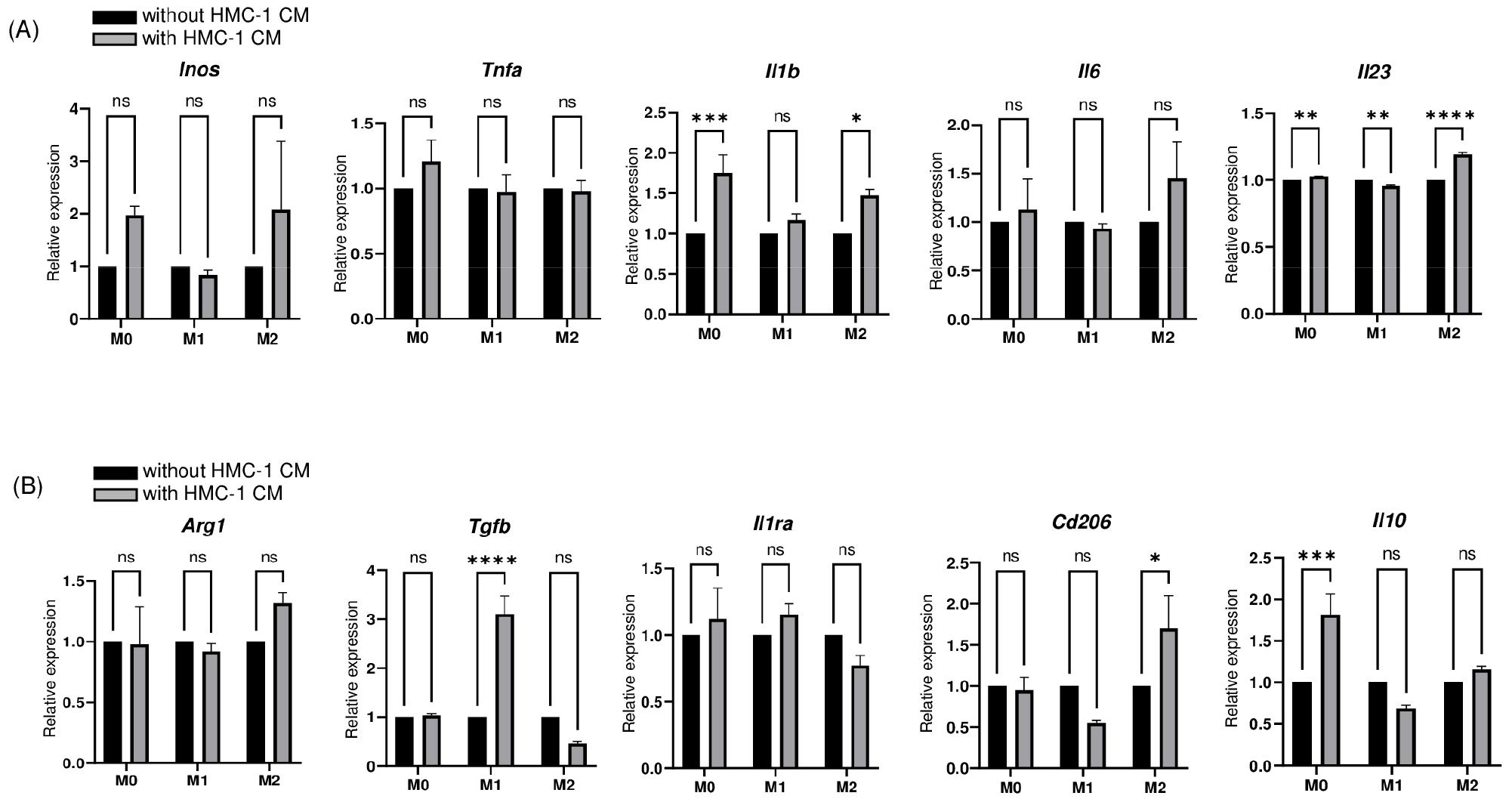
Fig. 3
M1-marker expression was modestly increased in M2-like macrophages induced by post-polarization treatment with HMC-1 CM. (A) qRT-PCR analysis of M1 markers, including Inos, Tnfa, Il1b, Il6, and Il23, in macrophages treated with or without HMC-1 CM after polarization into M1-like phenotype or M2-like phenotype. Data are presented as mean ± SEM and analyzed by two-way ANOVA with Sidak’s multiple comparison test (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001). (B) qRT-PCR analysis of M2 markers, including Arg1, Tgfb, Il1ra, Cd206, and Il10, in macrophages treated with or without HMC-1 CM after polarization into the M1-like phenotype or M2-like phenotype. Data are presented as mean ± SEM and analyzed by two-way ANOVA with Sidak’s multiple comparison test (*P<0.05, ***P<0.001, ****P<0.0001).
Significant induction of Tgfb and Il10 was observed in M1-like and M0 macrophages, respectively, upon exposure to HMC-1 CM. This suggests HMC-1 CM not only facilitated the plasticity of M2-like macrophage to M1-like macrophages but also maintained a modulatory influence, as indicated by the induction of Tgfb and Il10 expression. Thus, our results signify that HMC-1 CM enhances the plasticity of M2-like macrophages toward the M1-like phenotype. This transition, however, is subject to regulatory modulation, as demonstrated by the concurrent induction of Tgfb and Il10 expression.
Tryptase in HMC-1 CM mediates macrophage polarization to M1-like phenotype
Existing research has indicated the activation of protease‐activated receptor 2 (PAR2), a tryptase-binding receptor, promotes M1 macrophage polarization and inflammation in primary murine bone marrow-derived macrophages and the murine macrophage cell line RAW264.7 (11). Given MCs are primary sources of tryptase secretion, we hypothesized the M1-like macrophages polarization effect attributed to MCs could be mediated by tryptase. Therefore, we examined whether the inhibitory impact of HMC-1 CM on M1-like macrophages polarization could be mitigated by the addition of APC 366, a selective tryptase inhibitor (12).
Thus, APC 366 was co-administered with HMC-1 CM to M0 macrophages. The induction of M1 markers, specifically Inos, Tnfa, Il1b, and Il6 was reduced upon the addition of APC 366 to HMC-CM-treated macrophages (Fig. 4A). This effect was also observable in the context of Tgfb expression.
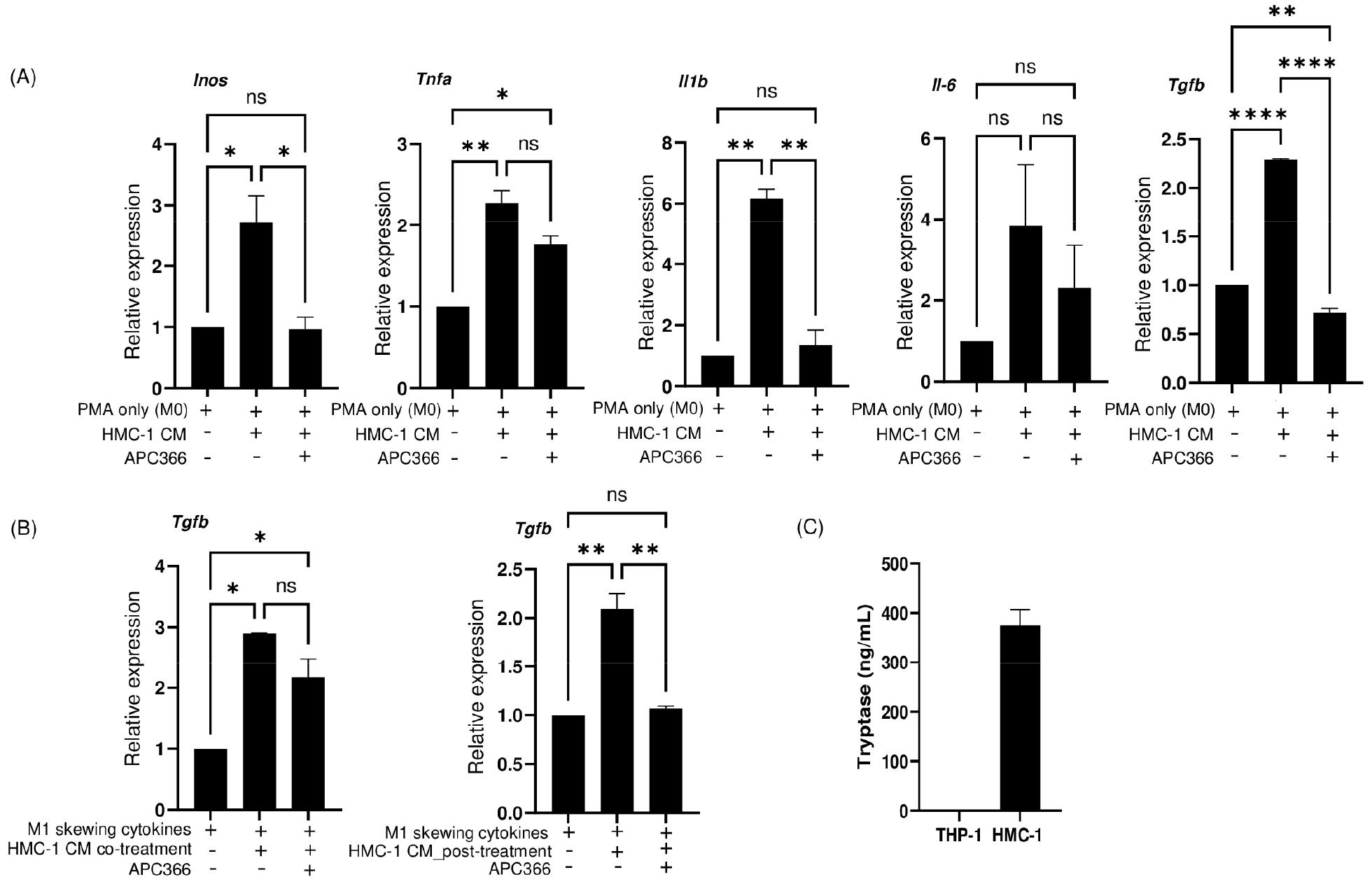
Fig. 4
Tryptase inhibition attenuates HMC-1 CM–induced gene expression. (A) qRT-PCR analysis of M1 markers, including Inos, Tnfa, Il1b, and Il6, and the M2 marker Tgfb in macrophages (M0 macrophages, M0 macrophages cultured with HMC-1 CM, and M0 macrophages cultured with HMC-1 CM and APC 366, a tryptase inhibitor). Data are presented as mean ± SEM analyzed by Ordinary one‑way ANOVA with Tukey’s multiple comparison test (*P<0.05, **P<0.01, ****P<0.0001). (B) qRT-PCR analysis of Tgfb in M1-like macrophages cultured under the indicated stimulation conditions. Data are presented as mean ± SEM analyzed by Ordinary one‑way ANOVA with Tukey’s multiple comparison test (*P<0.05, **P<0.01). (C) ELISA measurement of secreted tryptase in HMC-1 CM and THP-1 CM.
Furthermore, the influence of tryptase activity on the regulatory function of HMC-1 CM was examined by assessing the expression of Tgfb in M1-like macrophages (Fig. 4B). The increased expression of Tgfb induced by HMC-1 CM was evident both when introduced at the beginning of differentiation of M1-like macrophages and following differentiation. The effect of APC 366 was further confirmed in both cases where it was co-administered with HMC-1 CM. APC 366 treatment effectively counteracted the induction of Tgfb in M1-like macrophages in the presence of HMC-1 CM. This blocking effect was particularly significant when HMC-1 CM treatment occurred post-differentiation. The observation of high tryptase concentrations within HMC-1 CM further substantiates the involvement of tryptase in the effects associated with HMC-1 CM (Fig. 4C). Together, these findings underscore the potential involvement of tryptase in the regulatory interplay between HMC-1 CM and M1 polarization, further highlighting the complex mechanisms underlying immune cell modulation.
DISCUSSION
This study delves into the intricate interplay between MCs and macrophages, shedding light on the role of MCs in directing M0 macrophages toward the M1-like phenotype polarization. Significantly, we identified tryptase, a protease intrinsic to MCs, as a key mediator in this process. Our findings offer new insights into the regulatory dynamics of MCs within immune responses.
Upon activation, MCs play a critical role as secretory cells, releasing a diverse array of biologically active substances. These encompass pre-formed molecules stored in granules, such as histamine, TNF-α, heparin, lysosomal hydrolases, and proteases, allowing for rapid release upon activation. Additionally, MCs are capable of de novo synthesis of various mediators, including TNF-α, IL-6, IL-13, IL-1, IL-5, GM-CSF, leukotrienes, and prostaglandins (13). Importantly, MCs also engage in steady-state granule release under normal physiological conditions to maintain homeostasis by providing controlled amounts of bioactive compounds (14, 15, 16).
Macrophages are highly versatile phagocytic cells devoted to eliminating senescent or anomalous endogenous entities and recognizing and eliminating exogenous threats. They can adopt several functional phenotypes in response to microenvironmental cues, with extreme profiles being the inflammatory/killing phenotype (M1) and the anti-inflammatory/ healing phenotype (M2). During inflammation, M1 and M2 macrophages are activated alternately, and maintaining a stable balance between them is crucial for preserving homeostasis in the body.
If inflammation persists excessively, it can disrupt the balance between M1 and M2 macrophages, potentially leading to conditions such as autoimmune disorders, diabetes, allergies, atherosclerosis, obesity, and metabolic dysregulation (17). Thus, the imbalance between the two kinds of macrophages will have a significant impact on the illness process.
In the broader context of immune surveillance and homeostasis, MCs and macrophages represent sentinel cells that play pivotal roles in recognizing and responding to environmental signals. The coexistence of these two distinct myeloid-cell populations in all body tissues, especially barrier tissues, underscores a strategic adaptation by multicellular organisms to ensure rapid and adaptive responses to external challenges (16). Sentinel cells within barrier organs must balance robust pathogen defenses with controlled responses to prevent excessive inflammation. This delicate balance orchestrated by these sentinel cells is crucial for safeguarding overall health.
Our observations highlight the capacity of MCs to influence macrophage behavior, thus impacting the overall immune response. Specifically, THP-1 cells treated with PMA alone, termed M0 macrophages, exhibited elevated M1 marker expression when exposed to HMC-1 CM. Interestingly, HMC-1 CM also demonstrated a regulatory effect on M1-like macrophages by inducing the expression of Tgfb. This modulation was attenuated by a tryptase inhibitor, which likely targets tryptase present in HMC-1 CM, underscoring the crucial role of tryptase in mediating MC-induced M1-like phenotype polarization. This complex interaction between MCs and macrophages highlights the context-dependent synergy within immune modulation.
The interplay between MCs and macrophages is exemplified in type 2 immunity, characterized by the production of IL-4, IL-5, IL-9, and IL-13. This immune response is commonly observed in tissues during allergic inflammation or infection with helminth parasites (18). MCs promote type 2 immunity, thereby providing the environmental cytokine milieu leading to macrophage polarization to the M2 phenotype. Conversely, proinflammatory M1 macrophages and MCs are co-elevated in chronic adipose tissue inflammation, such as in obesity (19). These findings underscore the close interplay between MCs and macrophages in environments with varying stimuli.
The precise mechanisms underlying MC-macrophage interactions in normal physiological conditions remain unclear. Our findings demonstrate unstimulated MCs minimally influence polarization or the M1/M2 plasticity in macrophages exposed to sufficient stimulating factors. This implies unstimulated MCs exhibit limited secretion of essential factors such as INF-γ, IL-4, and IL-13, which are necessary for macrophage polarization. Our previous study indicated low basal levels of these secreted molecules in non-stimulated MCs under steady-state conditions (8). Thus, the impact of unstimulated MCs on macrophage polarization under sufficient stimulation is not synergistic.
We observed that HMC-1 CM robustly induced the expression of M1 marker genes rather than M2 maker genes in M0 macrophages stimulated with PMA only, particularly when treated at the beginning of the polarization culture period. However, this phenomenon was less pronounced when HMC-1 CM was added to M0 macrophages after 48 h of culture. These results suggest soluble factors released by MCs can potentiate polarization from M0 to M1. This process, however, required sustained stimulation over time. Furthermore, the introduction of HMC-1 CM resulted in increased Tgfb and Il10 expression in M0 or M1-like macrophages, regardless of the timing of administration. This dual functionality implies that while MCs typically induce M1 polarization to trigger an inflammatory response against rapidly invading pathogens if necessary, they concurrently foster appropriate anti-inflammatory responses to prevent excessive disruptions of homeostasis.
Our investigation delved into the factors driving MC-mediated M1-like phenotype polarization. We observed substantial secreted tryptase within MCs, aligning with steady-state granule release even under normal physiological conditions. Additionally, inhibition experiments revealed reduced effects of HMC-1 CM on M1-like phenotype polarization and anti-inflammatory gene expression in the presence of a tryptase inhibitor. Thus, MC-derived tryptase plays a pivotal role in promoting macrophage polarization possibly acting on protease-activated receptor 2 (PAR2), well known responding receptor to mast cell tryptase. It is reported that PAR2 promote macrophage polarization and inflammation via FOXO1 pathway (11). Also, PAR2 activate act through several molecular pathways such as nuclear factor kappa B (NF‐κB), mitogen‐activated protein kinase (MAPK/ERK) thereby increasing inflammatory cytokines (20, 21). The tryptase-dependent augmentation of macrophage polarization observed in our study is presumably mediated through the activation of intracellular signaling cascades via PAR2 stimulation, resulting in the transcriptional upregulation of pro-inflammatory cytokine genes. The tryptase inhibitor APC 366, we used in this study is a selective inhibitor of mast cell tryptase may vary depending on the target cell (22, 23, 24), further studies are needed to evaluate its effects in cells derived in vivo models or other cell lines.
In this study, our findings have deepened our understanding of the intricate crosstalk between MCs and macrophages, shedding light on their collaborative roles in immune responses. Additionally, we unveiled that MCs possess the capacity to influence macrophage polarization, particularly promoting the shift from the M0 to the M1 phenotype. The tryptase secreted by MCs emerged as a central mediator in orchestrating this regulatory effect. This novel insight into MC-mediated modulation of macrophage behavior opens avenues for future research aiming to decipher the intricate mechanisms governing immune responses. Furthermore, the enhanced wound healing observed in human epithelial cells under basal conditions due to MC-derived tryptase (25, 26) underscores the potency and potential therapeutic implications of this regulated interplay.
The elucidation of MC-mediated M1-like phenotype polarization and its dependence on tryptase offers potential insights into therapeutic interventions targeting immune dysregulation. The findings of this study underscore the significance of MCs as not only instigators of allergic and inflammatory responses but also as pivotal players in maintaining tissue homeostasis.
Also, mast cell tryptase-mediated macrophage polarization can significantly contribute to physiological functions. The interaction between eosinophils and macrophages has been well studied in the context of adaptive thermogenesis. IL-4 secreted by eosinophils induces M2 polarization of macrophages, supporting adaptive thermogenesis and maintaining tissue homeostasis (27, 28). Based on our findings, mast cell tryptase may activates macrophages, leading to the secretion of cytokines and chemokines that may recruit or activate eosinophils in adipose tissues. Furthermore, substances secreted by eosinophils could regulate macrophage polarization, supporting adaptive thermogenesis and adipose tissue metabolism.
Ultimately, the interaction between mast cells and macrophages is likely to play a critical role in various physiological and pathological conditions, such as obesity and metabolic disorders.
Considering that the substances produced by mast cells vary depending on the stimulus (29, 30), the role of MC would also differ in environments where M1 or M2 cells are predominant. For instance, MC produce various mediators, including IFN-β, which, either independently or in combination, can regulate the phagocytic activity of macrophages during the resolution of inflammation (31). Conversely, M2 cells and MC are key players in fibrotic diseases such as Dupuytren’s disease (32). Therefore, MCs are not only involved in the regulation of macrophages, but also play a significant role in the progression of pathological conditions through close interactions with macrophages in environments where specific macrophage phenotypes are predominant.
The identified role of tryptase as a key mediator prompts further investigation into its signaling pathways and downstream effects on macrophage polarization. Future studies might explore the broader implications of MC-derived tryptase, potentially uncovering its roles in other immune contexts and providing novel avenues for therapeutic manipulation. Moreover, the context-dependent nature of MC-macrophage interactions warrants exploration into how external stimuli modulate these responses and whether this interplay varies across tissues. Overall, these insights into the complex interplay between MCs and macrophages present promising avenues for advancing our understanding of immune regulation and its therapeutic potential.




