INTRODUCTION
Acinetobacter is a genus of gram-negative non-fermenting bacteria that are widely distributed in nature. The genus Acinetobacter is currently classified into more than 80 species (https://lpsn.dsmz.de/genus/acinetobacter). Acinetobacter species are a key source of infection in severely ill patients in the hospitals (1). Among the Acinetobacter species, A. baumannii is the most prevalent in clinical setting (2). In addition, non-baumanniiAcinetobacter species that are ubiquitous in the environment have evolved as important opportunistic pathogens for humans (3). In the prevalence of Acinetobacter species from 2022 to 2023 at long-term care facilities and general hospitals in 16 regions of Korea, 81% isolates were identified to A. baumannii, whereas the remaining isolates were non-baumannii species (4). Several non-baumanniiAcinetobacter species, including A. bereziniae, A. johnsonii, A. junii, A. lwoffii, A. nosocomialis, A. pitii, A. soli, and A. ursingii, are commonly isolated in clinical specimens (5, 6, 7).
A. baumannii is notorious for its ability to acquire resistance to antimicrobial agents, including cephalosporins, fluoroquinolones, and carbapenems, by intrinsic resistance and acquisition of resistance determinants (8). Carbapenem-resistant A. baumannii (CRAB)places top priority in the urgency of new antibiotic development by the World Health Organization (9). Although non-baumanniiAcinetobacter species are considered to be less resistant to antimicrobial agents than A. baumannii, multi-drug resistance (MDR), including carbapenem resistance, in non-baumanniiAcinetobacter species raises concern in clinical setting (3, 10, 11). Several Acinetobacter species intrinsically possess chromosomal carbapenem- hydrolyzing oxacillinases (CHDLs) such as blaOXA-51 for A. baumannii, blaOXA-23 for A. radioresistens, blaOXA-134 for A. lwoffii, blaOXA-211 for A. johnsonii, blaOXA-213 for A. calcoaceticus, and blaOXA-214 for A. haemolyticus(12). These species-specific oxacillinases have spread into other Acinetobacter species, for example exclusively existence of blaOXA-23 in Korean CRAB isolates (13). Ambler class B metallo-β-lactamases (MBLs), such as blaVIM and blaNDM, have been identified in non-baumanniiAcinetobacter species worldwide (11, 14, 15). Furthermore, coexistence of MBL and CHDL genes by transfer of MBL-carrying plasmids has been reported in non-baumanniiAcinetobacter species (14).
In Korea, non-baumanniiAcinetobacter species are increasingly prevalent in recent years. Three species, A. nosocomialis, A. seifertii, and A. pittii, are the most prevalent, although species distribution is different among the hospitals (10, 16). Clinical isolates of non-baumanniiAcinetobacter species are highly susceptible (>85%) to commonly used antimicrobial agents, including cephalosporins, β-lactams/β-lactamase inhibitors, fluoroquinolones, carbapenems, aminoglycosides, and colistin (17). However, resistance to these antimicrobial agents is gradually increasing in non-baumanniiAcinetobacter species from Korean hospitals (18). The present study investigated the species distribution, antimicrobial susceptibility, and carbapenem resistance mechanisms of non-baumanniiAcinetobacter species from three Korean hospitals.
MATERIALS AND METHODS
Bacterial strains and species identification
A total of 65 non-baumanniiAcinetobacter isolates collected in the Kyungpook National University Hospital-National Culture Collection for Pathogens (KNUH-NCCP), Gyeongsang National University Hospital-NCCP (GNUH-NCCP), and Jeonbuk National University Hospital-NCCP (JNUH-NCCP) between 2017 and 2020 were used in this study. Species was initially identified by the VITEK 2 system (bioMérieux, Marcy-l’Étoile, France), and then identified to each Acinetobacter species by the matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry analysis (Bruker Daltonics, Bremen, Germany) and sequencing of the 16S rDNA and rpoB genes (19).
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of non-baumanniiAcinetobacter isolates was determined using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (20). Fifteen antimicrobial agents were used in this study: aminoglycosides (amikacin, gentamicin, and tobramycin), antipseudomonal penicillins (piperacillin), penicillins/β-lactamase inhibitors (ampicillin/sulbactam, piperacillin/tazobactam, ticarcillin/clavulanic acid), carbapenems (imipenem and meropenem), cephalosporins (ceftazidime), folic acid pathway inhibitors (trimethoprim/ sulfamethoxazole), fluoroquinolones (ciprofloxacin), tetracycline derivatives (minocycline and tigecycline), and polymyxins (colistin). Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains. The susceptibility to tigecycline was determined by the criteria of the US Food and Drug Administration for Enterobacteriaceae (21). The MDR or extensively-drug resistant (XDR) phenotypes were determined according to the criteria proposed by Magiorakos et al (22).
Screening of carbapenemase production and amplification of resistance genes
The production of carbapenemases in carbapenem resistant non-baumanniiAcinetobacter isolates was determined using the Rapidec Carba NP test (bioMérieux) (23). Polymerase chain reaction was conducted to detect carbapenem resistance determinants. DNA templates were prepared by boiling of bacteria for 10 min after bacteria were grown in lysogeny broth (LB) overnight. To detect carbapenemase genes in the Rapidec Carba NP test-positive bacterial isolates, primers specific for blaIMP, blaNDM, blaVIM, blaOXA-23-like, blaOXA-58, blaOXA-211, and blaOXA-213 were used (Table 1) (24, 25, 26, 27, 28, 29). The full lengths of blaOXA, blaNDM, and blaVIM genes were amplified and sequenced to identify the subtypes of genes.
Table 1.
Primers used in this study
| Primers | Sequence (5' to 3') | Genes | Use | References |
|---|---|---|---|---|
| blaIMP-F | GAATAGRRTGGCTTAAYTCTC | blaIMP | Detection | (24) |
| blaIMP-R | CCAAACYACTASGTTATC | |||
| blaNDM-F | TTGGCCTTGCTGTCCTTG | blaNDM | Detection | (25) |
| blaNDM-R | ACACCAGTGACAATATCACCG | |||
| blaSPM-F | CTGCTTGGATTCATGGGCGC | blaSPM | Detection | (26) |
| blaSPM-R | CCTTTTCCGCGACCTTGATC | |||
| blaVIM-F | GTTTGGTCGCATATCGCAAC | blaVIM | Detection | (26) |
| blaVIM-R | AATGCGCAGCACCAGGATAG | |||
| blaoxa-48-F | TTGGTGGCATCGATTATCGG | blaoxa-48 | Detection | (26) |
| blaoxa-48-R | GAGCACTTCTTTTGTGATGGC | |||
| blaoxa-58-F | CGATCAGAATGTTCAAGCGC | blaoxa-58 | Detection | (26) |
| blaoxa58-R | ACGATTCTCCCCTCTGCGC | |||
| blaoxa-23-F | ATGAATAAATATTTTACTTGCTATG | blaoxa-23-like | Detection | This study |
| blaoxa-23-R | TTAAATAATATTCAGCTGTT | |||
| blaoxa-211-F | ATGAAAAATTTACAGTTGGGCC | blaoxa-211-like | Detection | This study |
| blaoxa-211-R | TTAAATTATCCCCAGTGCTG | |||
| blaoxa-213-F | ATGACTAAAAAAGCTCTTTTCTTTGC | blaoxa-213-like | Detection | This study |
| blaoxa-213-R | TTATAAAATACCTAGCTGCTCTAATCC | |||
| ISAba1-F | AAGCACTTGATGGGCAAGGC | ISAba1 | Detection | (27) |
| blaOXA-23 r | CGGGATCCCGTTAAATAATATTCAGGTC | |||
| ISAba3-F | CAATCAAATGTCCAACCTGC | ISAba3 | Amplification | This study |
| blaOXA-58 r | TTATAAATAATGAAAAACACC | |||
| blaNDM-Full-F | GTTTTCCCAGTCACGACGTTATGGAATTGCCCAATATTATGCACCCGGTCG | blaNDM-1 | Amplification and sequencing | (28) |
| blaNDM-Full-R | CAGGAAACAGCTATGACCATGATCAGCGCAGCTTGTCGGCCATG | |||
| blaVIM-Full-F | GTTTTCCCAGTCACGACGTTATGTTCAAACTTTTGAGTAAG | blaVIM | Amplification and sequencing | (28, 29) |
| blaVIM-Full-R | CAGGAAACAGCTATGACCATGACTACTCAACGACTGAGCGAT |
RESULTS
Isolation of non-baumannii Acinetobacter isolates
Non-baumanniiAcinetobacter isolates (n = 65) were collected from KNUH-NCCP (n = 19), GNUH-NCCP (n = 23), and JNUH-NCCP (n = 23) between 2017 and 2020. The isolates were from 2017 (n = 8), 2018 (n = 12), 2019 (n = 12), and 2020 (n = 33). Non-baumanniiAcinetobacter isolates were obtained from sputum and respiratory tract (n = 17), urine (n = 17), blood (n = 15), pus (n = 9), body fluids (n = 4), vaginal discharge (n = 2), and skin tissue (n = 1).
Species distribution of non-baumannii Acinetobacter isolates
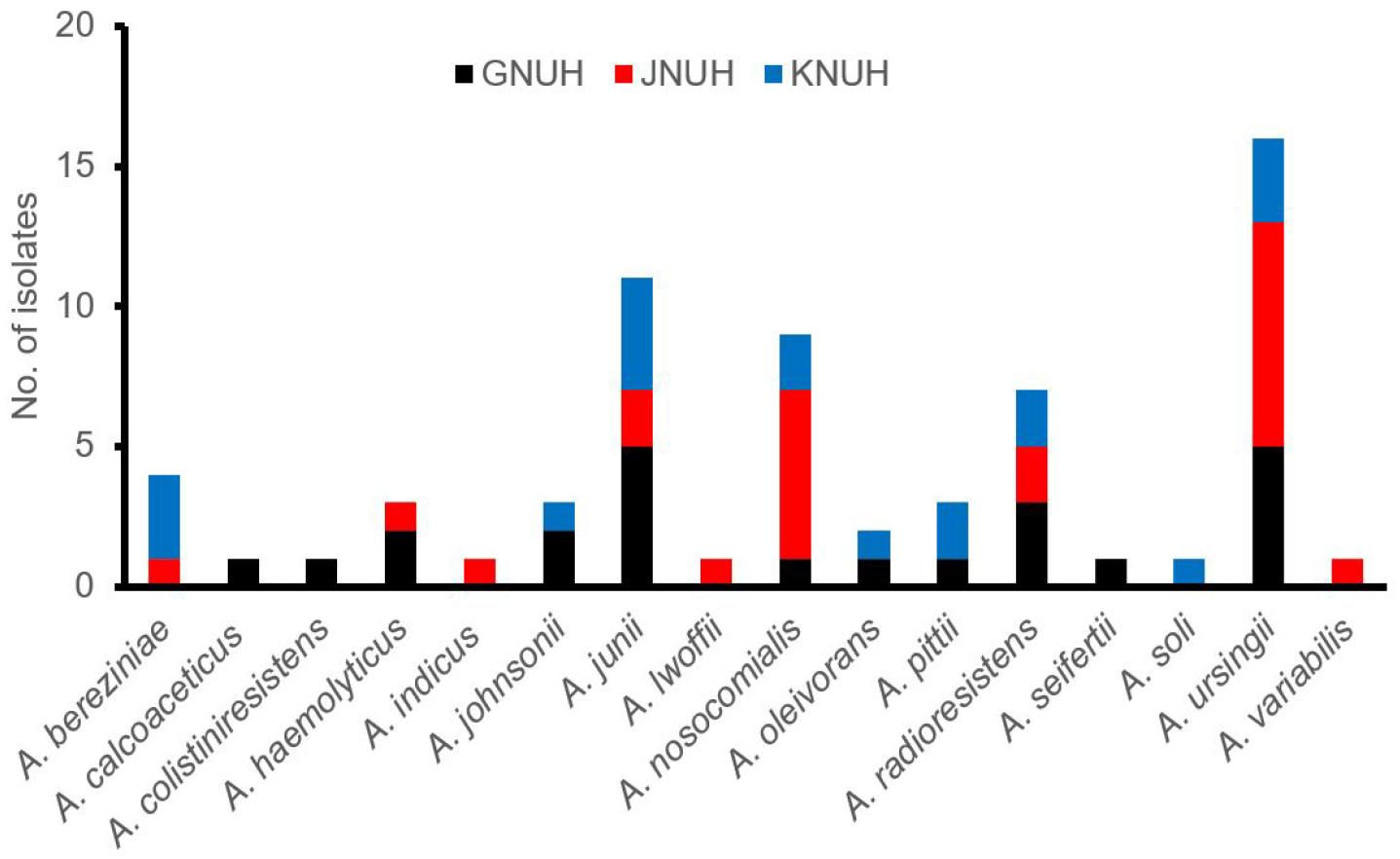
A total of 16 different species were identified among the 65 non-baumanniiAcinetobacter isolates using the MALDI-TOF mass spectrometry and 16S rDNA and rpoB sequencing (Fig. 1). Four species, including A. ursingii (n = 16), A. junii (n = 11), A. nosocomialis (n = 9), and A. radioresistens (n = 7),were prevalent, accounting for 66% of the isolates. These four species were detected in three hospitals. Seven species, including A. calcoaceticus, Acinetobacter genomosp. 13BJ, A. indicus, A. lwoffii, A. seifertii, A. soli, and A. variabilis, were detected in each isolate.
Antimicrobial susceptibility of non-baumannii Acinetobacter isolates
Sixty-five non-baumanniiAcinetobacter isolates exhibited less than 30% resistance rates to all tested antimicrobial agents. Resistance to piperacillin (26.2%), ciprofloxacin (23.1%), gentamicin (23.1%), and ceftazidime (21.5%) was a relatively high, whereas no isolates were resistant to minocycline and tigecycline (Table 2). Resistance rates to the remaining nine antimicrobial agents were ranged from 6.2% (amikacin) ~ 18.5% (tobramycin and piperacillin/tazobactam). Thirty-three (50.8%) isolates were susceptible to all tested antimicrobial agents. The MDR and XDR phenotypes were identified in 11 non-baumanniiAcinetobacter isolates and one A. bereziniae isolate, respectively. Twenty (30.8%) isolates were resistant to one or two antimicrobial classes.
Table 2.
Antimicrobial susceptibility of non-baumanniiAcinetobacter isolates
| No. of resistant isolates | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | AMK | GEN | TOB | PIP | SAM | TZP | TCC | IPM | MEM | CAZ | SXT | CIP | CL | MIN | TGC |
| A. bereziniae (n=4) | 3 | 3 | 3 | 3 | 1 | 2 | 2 | 1 | 1 | 3 | 2 | 3 | 2 | 0 | 0 |
| A. calcoaceticus (n =1) | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| Acinetobacter genomosp. 13BJ(n=1) | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. haemolyticus (n=3) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. indicus (n=1) | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| A. johnsonii (n=3) | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
| A. junii (n=11) | 0 | 2 | 1 | 3 | 1 | 2 | 2 | 2 | 2 | 3 | 1 | 1 | 2 | 0 | 0 |
| A. lwoffii (n=1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. nosocomialis (n=9) | 0 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 4 | 0 | 0 | 0 |
| A. oleivorans (n=2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| A. pitii (n=3) | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| A. radioresistens (n=7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| A. soli (n=1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| A. seifertii (n=1) | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. ursingii (n=16) | 0 | 2 | 1 | 3 | 1 | 2 | 2 | 0 | 0 | 4 | 1 | 3 | 0 | 0 | 0 |
| A. variabilis (n=1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (n=65) | 4 (6.2) | 15 (23.1) | 12 (18.5) | 17 (26.2) | 8 (12.3) | 12 (18.5) | 9 (13.8) | 7 (10.8) | 7 (10.8) | 14 (21.5) | 8 (12.3) | 15 (23.1) | 6 (9.2) | 0 (0) | 0 (0) |
Abbreviations. AMK, amikacin; GEN, gentamicin; TOB, tobramycin; PIP, piperacillin; SAM, ampicillin/sulbactam; TZP, piperacillin/Tazobactam; TCC, ticarcillin/clavulanic acid; IPM, imipenem; MEM, meropenem; FEP, cefepime; CAZ, ceftazidime; SXT, trimethoprim/sulfamethoxazole; CIP, ciprofloxacin; CL, colistin; MIN, minocycline; TGC, tigecycline.
Carbapenem resistance of non-baumannii Acinetobacter isolates
Seven non-baumanniiAcinetobacter isolates, including A. bereziniae (n = 1), A. calcoaceticus (n = 1), A. johnsonii (n = 1), A. junii (n = 2), A. nosocomialis (n = 1), and A. pittii (n = 1), were resistant to imipenem and meropenem. The Rapidec Carba NP test revealed that all seven isolates produced carbapenemases. A total of six different carbapenemase genes were detected in seven non-baumanniiAcinetobacter isolates: blaOXA-23, blaOXA-58, blaOXA-211, and blaOXA-213 for class D oxacillinases genes and blaNDM-1 and blaVIM-2 for class B MBL genes (Table 3). A. calcoaceticus isolate carried three different carbapenemase genes, blaOXA-58, blaOXA-213, and blaNDM-1. A. pittii isolate carried two different carbapenemase genes, blaOXA-23, and blaOXA-213. One A. junii isolate carried two MBL genes, blaVIM-2 and blaNDM-1. blaOXA-23 and blaOXA-58 genes carried ISAba1 and ISAba3 promoters, respectively. However, A. johnsonii isolate did not carry any carbapenemase genes tested.
Table 3.
Carbapenem resistance genes identified in non-baumanniiAcinetobacter isolates
DISCUSSION
This study characterized 65 clinical non-baumanniiAcinetobacter isolates collected from the patients admitted to three hospitals located in southern part of South Korea over four years. These isolates were identified to 16 different non-baumanniiAcinetobacter species using the MALDI-TOF mass spectrometry and 16S rDNA and rpoB sequencing. Of the identified non-baumanniiAcinetobacter species, A. ursingii (24.6%) was the most prevalent, followed by A. junii (16.9%), A. nosocomialis (13.8%), and A. radioresistens (10.8%). These prevalent species were detected in three study hospitals and the high risk group of patients, such as patients admitted to intensive care units, might be exposed to non-baumannii Acinetobacter. A. pittii and A. nosocomialis that belonged to A. calcoaceticus-A. baumannii complex have been known to the most prevalent among non-baumanniiAcinetobacter species (30, 31), but only three A. pittii isolates were detected in two hospitals. A. baumannii is the most frequently isolated from sputum or respiratory tract, but rarely isolated from urinary tract (8, (13). However, in this study, 17 of 65 non-baumanniiAcinetobacter isolates were obtained from urine sample. These results suggest that non-baumanniiAcinetobacter species commonly infect urinary tract as well as respiratory tract.
A half of non-baumanniiAcinetobacter isolates were susceptible to all tested antimicrobial agents. Of the 32 drug-resistant isolates, 20 were resistant to one or two antimicrobial agents and 11 exhibited the MDR phenotype, which was resistant to three antimicrobial classes. Only one A. bereziniae isolate exhibited XDR phenotype, which was susceptible to minocycline and tigecycline. Resistance rates to tested antimicrobial agents did not exceed 25%. Antimicrobial resistance in non-baumanniiAcinetobacter isolates from KNUH is significantly lower than A. baumannii isolates from the same hospital during the study period (13). More than 95% of A. baumannii isolates exhibited MDR and XDR phenotypes (32). These results suggest that most clinical non-baumanniiAcinetobacter isolates are susceptible to commonly used antimicrobial agents, unlike clinical A. baumannii isolates.
Of the 65 non-baumanniiAcinetobacter isolates, seven (10.8%) were resistant to carbapenems. blaOXA-211 and blaOXA-213 are species-specific intrinsic CHDLs for A. johnsonii and A. calcoaceticus, respectively (12). In addition, blaOXA-23 identified in A. nosocomialis and A. pittii, blaOXA-58 in A. calcoaceticus, blaVIM-2 in A. junii, and blaNDM-1 in A. junii and A. calcoaceticus were acquired carbapenemase genes. CRAB isolates from Korean hospitals exclusively carried blaOXA-23 regardless of clonal linages (13, 18), suggesting the transfer of blaOXA-23 from CRAB to A. nosocomialis and A. pittii. blaOXA-58 was not detected in Korean CRAB isolates in recent years, but this gene was detected in non-baumanniiAcinetobacter species, including A. bereziniae, A. junii,A. nosocomialis, and A. pittii from other countries (33, 34, 35, 36). MBL genes such as blaIMP-1 and blaNDM-1 were also detected in non-baumanniiAcinetobacter species, often in combination with class D carbapenemase genes worldwide (34, 36, 37, 38). In Korea, three MBL genes, blaIMP-1, blaNDM-1, and blaSIM-1, were detected in non-baumanniiAcinetobacter isolates (10, 11, 16). blaIMP-1 was the most prevalent, followed by blaVIM-2 and blaSIM-1 in non-baumanniiAcinetobacter species from 2005 to 2012 (9). However, blaIMP-1 and blaSIM-1 were not detected in this study. blaNDM-1 was detected in A. pittii isolates from Korean hospitals located in Seoul and Daejeon (10, 16). In this study, blaNDM-1 was detected in one A. calcoaceticus and two A. junii isolates. Moreover, A. junii isolate co-carried tow MBL genes, blaVIM-2 and blaNDM-1. The genetic background of carbapenem resistance in non-baumanniiAcinetobacter species should be further analyzed. Because non-baumanniiAcinetobacter species play a role in potential reservoir of carbapenem resistance genes (39), the spread of carbapenem resistance genes should be carefully monitored among Acinetobacter species.
AUTHOR CONTRIBUTIONS
Conceptualization, J.C.L. and S.K.; Data curation; J.C.L. and S.K.; Formal analysis, J.C.L. and S.K.; Funding acquisition, S.K.; Investigation, S.K. and M.S.R.; Methodology, S.K., M.S.R., B.K., S.-Y.K., and N.K.; Project administration, S.K. and J.C.L.; Resources, J.C.L. and D.E.L.; Supervision, J.C.L.; Visualization, S.K. and J.C.L.; Roles/Writing - original draft, S.K. and J.C.L.; and Writing - review & editing, J.C.L.